Abstract
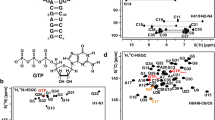
RNA aptamers are used in a wide range of biotechnological or biomedical applications. In many cases the high resolution structures of these aptamers in their ligand-complexes have revealed fundamental aspects of RNA folding and RNA small molecule interactions. Fluorescent RNA-ligand complexes in particular find applications as optical sensors or as endogenous fluorescent tags for RNA tracking in vivo. Structures of RNA aptamers and aptamer ligand complexes constitute the starting point for rational function directed optimization approaches. Here, we present the NMR resonance assignment of an RNA aptamer binding to the fluorescent ligand tetramethylrhodamine (TMR) in complex with the ligand 5-carboxy-tetramethylrhodamine (5-TAMRA) as a starting point for a high-resolution structure determination using NMR spectroscopy in solution.


Similar content being viewed by others
References
Carothers JM, Goler JA, Kapoor Y, Lara L, Keasling JD (2010) Selecting RNA aptamers for synthetic biology: investigating magnesium dependence and predicting binding affinity. Nucl Acids Res 38:2736–2747
Dallmann A, Simon B, Duszczyk MM, Kooshapur H, Pardi A, Bermel W, Sattler M (2013) Efficient detection of hydrogen bonds in dynamic regions of RNA by sensitivity-optimized NMR pulse sequences. Angew Chem Int Ed Engl 52:10487–10490
Dingley AJ, Grzesiek S (1998) Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide (2)J(NN) couplings. J Am Chem Soc 120:8293–8297
Duchardt-Ferner E, Weigand JE, Ohlenschläger O, Schmidtke SR, Suess B, Wöhnert J (2010) Highly modular structure and ligand binding by conformational capture in a minimalistic riboswitch. Angew Chem Int Ed Engl 49:6216–6219
Duchardt-Ferner E, Gottstein-Schmidtke SR, Weigand JE, Ohlenschläger O, Wurm JP, Hammann C, Suess B, Wöhnert J (2016) What a difference an OH makes: conformational dynamics as the basis for the ligand specificity of the neomycin-sensing riboswitch. Angew Chem Int Ed Engl 55:1527–1530
Ellington AD, Szostak JW (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346:818–822
Fiala R, Jiang F, Patel DJ (1996) Direct correlation of exchangeable and nonexchangeable protons on purine bases in C-13, N-15-labeled RNA using a HCCNH-TOCSY experiment. J Am Chem Soc 118:689–690
Fiala R, Jiang F, Sklenar V (1998) Sensitivity optimized HCN and HCNCH experiments for C-13/N-15 labeled oligonucleotides. J Biomol NMR 12:373–383
Fürtig B, Richter C, Bermel W, Schwalbe H (2004) New NMR experiments for RNA nucleobase resonance assignment and chemical shift analysis of an RNA UUCG tetraloop. J Biomol NMR 28:69–79
Hermann T, Patel DJ (2000) Adaptive recognition by nucleic acid aptamers. Science 287:820–825
Heus HA, Pardi A (1991) Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science 253:191–194
Jiang F, Kumar RA, Jones RA, Patel DJ (1996) Structural basis of RNA folding and recognition in an AMP-RNA aptamer complex. Nature 382:183–186
Legault P, Farmer BT, Mueller L, Pardi A (1994) Through-bond correlation of adenine protons in a C-13-labeled ribozyme. J Am Chem Soc 116:2203–2204
Majumdar A, Kettani A, Skripkin E (1999) Observation and measurement of internucleotide 2JNN coupling constants between 15 N nuclei with widely separated chemical shifts. J Biomol NMR 14:67–70
Marino JP, Schwalbe H, Anklin C, Bermel W, Crothers DM, Griesinger C (1994) A 3-dimensional triple-resonance H-1, C-13, P-31 experiment—sequential through-bond correlation of ribose protons and intervening phosphorus along the RNA oligonucleotide backbone. J Am Chem Soc 116:6472–6473
Marino JP, Diener JL, Moore PB, Griesinger C (1997) Multiple-quantum coherence dramatically enhances the sensitivity of CH and CH2 correlations in uniformly C-13-labeled RNA. J Am Chem Soc 119:7361–7366
Milligan JF, Uhlenbeck OC (1989) Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol 180:51–62
Nikonowicz EP, Pardi A (1992) Three-dimensional heteronuclear NMR studies of RNA. Nature 355:184–186
Ohlenschläger O, Wöhnert J, Bucci E, Seitz S, Häfner S, Ramachandran R, Zell R, Görlach M (2004) The structure of the stemloop D subdomain of coxsackievirus B3 cloverleaf RNA and its interaction with the proteinase 3C. Structure 12:237–248
Phan AT (2000) Long-range imino proton-13C J-couplings and the through-bond correlation of imino and non-exchangeable protons in unlabeled DNA. J Biomol NMR 16:175–178
Piotto M, Saudek V, Sklenar V (1992) Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR 2:661–665
Simon B, Zanier K, Sattler M (2001) A TROSY relayed HCCH-COSY experiment for correlating adenine H2/H8 resonances in uniformly 13C-labeled RNA molecules. J Biomol NMR 20:173–176
Sklenar V, Brooks BR, Zon G, Bax A (1987) Absorption mode two-dimensional NOE spectroscopy of exchangeable protons in oligonucleotides. FEBS Lett 216:249–252
Sklenar V, Peterson RD, Rejante MR, Feigon J (1993) 2-Dimensional and 3-dimensional HCN experiments for correlating base and sugar resonances in N-15, C-13-labeled RNA oligonucleotides. J Biomol NMR 3:721–727
Sklenar V, Peterson RD, Rejante MR, Feigon J (1994) Correlation of nucleotide base and sugar protons in a 15 N-labeled HIV-1 RNA oligonucleotide by 1H-15N HSQC experiments. J Biomol NMR 4:117–122
Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249:505–510
Wijmenga SS, van Buuren BNM (1998) The use of NMR methods for conformational studies of nucleic acids. Prog Nucl Mag Res Spectrosc 32:287–387
Wöhnert J, Dingley AJ, Stoldt M, Görlach M, Grzesiek S, Brown LR (1999a) Direct identification of NH…N hydrogen bonds in non-canonical base pairs of RNA by NMR spectroscopy. Nucl Acids Res 27:3104–3110
Wöhnert J, Ramachandran R, Görlach M, Brown LR (1999b) Triple-resonance experiments for correlation of H5 and exchangeable pyrimidine base hydrogens in (13)C, (15)N-labeled RNA. J Magn Reson 139:430–433
Wunderlich CH, Spitzer R, Santner T, Fauster K, Tollinger M, Kreutz C (2012) Synthesis of (6-C-13)pyrimidine nucleotides as spin-labels for RNA dynamics. J Am Chem Soc 134:7558–7569
Yang Y, Kochoyan M, Burgstaller P, Westhof E, Famulok M (1996) Structural basis of ligand discrimination by two related RNA aptamers resolved by NMR spectroscopy. Science 272:1343–1347
Zimmermann GR, Jenison RD, Wick CL, Simorre JP, Pardi A (1997) Interlocking structural motifs mediate molecular discrimination by a theophylline-binding RNA. Nat Struct Biol 4:644–649
Acknowledgments
This work was supported by the Adolf-Messer Foundation, the Center for Biomagnetic Resonance (BMRZ) of the Goethe-University Frankfurt and the Deutsche Forschungsgemeinschaft (DFG) (SFB 902 “Molecular principles of RNA-based regulation” B10) and the Austrian Science Fund (FWF, projects P28725 and P 26550).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Duchardt-Ferner, E., Juen, M., Kreutz, C. et al. NMR resonance assignments for the tetramethylrhodamine binding RNA aptamer 3 in complex with the ligand 5-carboxy-tetramethylrhodamine. Biomol NMR Assign 11, 29–34 (2017). https://doi.org/10.1007/s12104-016-9715-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-016-9715-6