Abstract
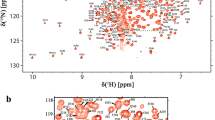
Rad23 functions in nucleotide excision repair and proteasome-mediated protein degradation. It has four distinct structural domains that are connected by flexible linker regions, including an N-terminal ubiquitin-like (UBL) domain that binds proteasomes. We report in this NMR study the 1H, 15N and 13C resonance assignments for the backbone and side chain atoms of the Rad23 UBL domain (Rad23UBL) with BioMagResBank accession number 25825. We find that a Rad23 proline amino acid (P20) located in a loop undergoes isomerization. The secondary structural elements predicted from the NMR data fit well to that of the Rad23UBL when complexed with E4 ubiquitin ligase Ufd2, as reported in a crystallographic structure. These complete assignments can be used to study the protein dynamics of the Rad23UBL and its interaction of with other ubiquitin receptors or proteasome subunits.



Similar content being viewed by others
References
Bartels C, Xia T-H, Billeter M, Güntert P, Wüthrich K (1995) The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J Biomol NMR 6:1–10
Bertolaet BL, Clarke DJ, Wolff M, Watson MH, Henze M, Divita G, Reed SI (2001) UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat Struct Biol 8:417–422. doi:10.1038/87575
Dantuma NP, Heinen C, Hoogstraten D (2009) The ubiquitin receptor Rad23: at the crossroads of nucleotide excision repair and proteasomal degradation. DNA Repair (Amst) 8:449–460. doi:10.1016/j.dnarep.2009.01.005
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Elsasser S et al (2002) Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol 4:725–730. doi:10.1038/ncb845
Fujiwara K et al (2004) Structure of the ubiquitin-interacting motif of S5a bound to the ubiquitin-like domain of HR23B. J Biol Chem 279:4760–4767. doi:10.1074/jbc.M309448200
Gomez TA, Kolawa N, Gee M, Sweredoski MJ, Deshaies RJ (2011) Identification of a functional docking site in the Rpn1 LRR domain for the UBA-UBL domain protein Ddi1. BMC Biol 9:33. doi:10.1186/1741-7007-9-33
Guzder SN, Sung P, Prakash L, Prakash S (1998) Affinity of yeast nucleotide excision repair factor 2, consisting of the Rad4 and Rad23 proteins, for ultraviolet damaged DNA. J Biol Chem 273:31541–31546
Hanzelmann P, Stingele J, Hofmann K, Schindelin H, Raasi S (2010) The yeast E4 ubiquitin ligase Ufd2 interacts with the ubiquitin-like domains of Rad23 and Dsk2 via a novel and distinct ubiquitin-like binding domain. J Biol Chem 285:20390–20398. doi:10.1074/jbc.M110.112532
Hiyama H et al (1999) Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. J Biol Chem 274:28019–28025
Husnjak K et al (2008) Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453:481–488. doi:10.1038/nature06926
Kang Y, Vossler RA, Diaz-Martinez LA, Winter NS, Clarke DJ, Walters KJ (2006) UBL/UBA ubiquitin receptor proteins bind a common tetraubiquitin chain. J Mol Biol 356:1027–1035. doi:10.1016/j.jmb.2005.12.001
Kang Y, Chen X, Lary JW, Cole JL, Walters KJ (2007a) Defining how ubiquitin receptors hHR23a and S5a bind polyubiquitin. J Mol Biol 369:168–176. doi:10.1016/j.jmb.2007.03.008
Kang Y, Zhang N, Koepp DM, Walters KJ (2007b) Ubiquitin receptor proteins hHR23a and hPLIC2 interact. J Mol Biol 365:1093–1101. doi:10.1016/j.jmb.2006.10.056
Mueller TD, Feigon J (2003) Structural determinants for the binding of ubiquitin-like domains to the proteasome. EMBO J 22:4634–4645. doi:10.1093/emboj/cdg467
Rosenzweig R, Bronner V, Zhang D, Fushman D, Glickman MH (2012) Rpn1 and Rpn2 coordinate ubiquitin processing factors at proteasome. J Biol Chem 287:14659–14671. doi:10.1074/jbc.M111.316323
Ryu KS, Lee KJ, Bae SH, Kim BK, Kim KA, Choi BS (2003) Binding surface mapping of intra- and interdomain interactions among hHR23B, ubiquitin, and polyubiquitin binding site 2 of S5a. J Biol Chem 278:36621–36627. doi:10.1074/jbc.M304628200
Shen Y, Delaglio F, Cornilescu G, Bax A (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR 44:213–223. doi:10.1007/s10858-009-9333-z
Shi Y et al (2016) Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 351:831. doi:10.1126/science.aad9421
Walters KJ, Lech PJ, Goh AM, Wang Q, Howley PM (2003) DNA-repair protein hHR23a alters its protein structure upon binding proteasomal subunit S5a. Proc Natl Acad Sci USA 100:12694–12699. doi:10.1073/pnas.1634989100
Wang Q, Goh AM, Howley PM, Walters KJ (2003) Ubiquitin recognition by the DNA repair protein hHR23a. Biochemistry 42:13529–13535. doi:10.1021/bi035391j
Wishart DS, Sykes BD (1994) The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR 4:171–180
Acknowledgments
This research was funded by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research to K.J.W. We thank Janusz Koscielniak for his technical assistance with the NMR facility.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, X., Walters, K.J. 1H, 15N, 13C resonance assignments for Saccharomyces cerevisiae Rad23 UBL domain. Biomol NMR Assign 10, 291–295 (2016). https://doi.org/10.1007/s12104-016-9686-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-016-9686-7