Abstract
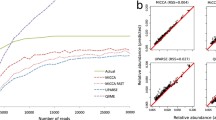
Taxonomic profiling, using hyper-variable regions of 16S rRNA, is one of the important goals in metagenomics analysis. Operational taxonomic unit (OTU) clustering algorithms are the important tools to perform taxonomic profiling by grouping 16S rRNA sequence reads into OTU clusters. Presently various OTU clustering algorithms are available within different pipelines, even some pipelines have implemented more than one clustering algorithms, but there is less literature available for the relative performance and features of these algorithms. This makes the choice of using these methods unclear. In this study five current state-of-the-art OTU clustering algorithms (CDHIT, Mothur’s Average Neighbour, SUMACLUST, Swarm, and UCLUST) have been comprehensively evaluated on the metagenomics sequencing data. It was found that in all the datasets, Mothur’s average neighbour and Swarm created more number of OTU clusters. Based on normalized mutual information (NMI) and normalized information difference (NID), Swarm and Mothur’s average neighbour showed better clustering qualities than others. But in terms of time complexity the greedy algorithms (SUMACLUST, CDHIT, and UCLUST) performed well. So there is a trade-off between quality and time, and it is necessary while analysing large size of 16S rRNA gene sequencing data.



Similar content being viewed by others
References
Albanese D, Fontana P, De Filippo C, Cavalieri D and Donati C 2015 MICCA: A complete and accurate software for taxonomic profiling of metagenomic data. Sci. Rep. 5 1–7
Allali I, Arnold JW, Roach J, et al. 2017 A comparison of sequencing platforms and bioinformatics pipelines for compositional analysis of the gut microbiome. BMC Microbiol. 17 194
Altschul SF, Gish W, Miller W, Myers EW and Lipman DJ 1990 Basic local alignment search tool. J. Mol. Biol. 215 403–410
Angly FE, Willner D, Rohwer F, Hugenholtz P and Tyson GW 2012 Grinder: A versatile amplicon and shotgun sequence simulator. Nucleic Acids Res. 40 e94
Bernard G, Pathmanathan JS, Lannes R, Lopez P and Bapteste E 2018 Microbial dark matter investigations: how microbial studies transform biological knowledge and empirically sketch a logic of scientific discovery. Genome Biol. Evol. 10 707–715
Bhat AH and Prabhu P 2017 OTU clustering: A window to analyse uncultured microbial world. Int. J. Sci. Res. Comput. Sci. Eng. 5 62–68
Bleidorn C 2016 Third generation sequencing: Technology and its potential impact on evolutionary biodiversity research. Syst. Biodivers 14 1–8
Cai Y and Sun Y 2011 ESPRIT-Tree: Hierarchical clustering analysis of millions of 16S rRNA pyrosequences in quasilinear computational time. Nucleic Acids Res. 39 1–10
Caporaso JG, Kuczynski J, Stombaugh J, et al. 2010 QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7 335–336
Chen W, Zhang CK, Cheng Y, Zhang S and Zhao H 2013 A Comparison of methods for clustering 16S rRNA sequences into OTUs. PLoS One 8 e70837
Clemente JC, Ursell LK, Parfrey LW and Knight R 2012 The impact of the gut microbiota on human health: An Integrative view. Cell 148 1258–1270
Edgar RC 2004 MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 1792–1797
Edgar RC 2010 Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26 2460–2461
Ghodsi M, Liu B and Pop M 2011 DNACLUST: Accurate and efficient clustering of phylogenetic marker genes. BMC Bioinformatics 12 271
Giongo A, Davis-richardson AG, Crabb DB and Triplett EW 2010 TaxCollector: Modifying current 16S rRNA databases for the rapid classification at six taxonomic levels. Diversity 2 1015–1025
Hao X, Jiang R and Chen T 2011 Clustering 16S rRNA for OTU prediction: A method of unsupervised Bayesian clustering. Bioinformatics 27 611–618
Janda JM and Abbott SL 2007 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J. Clin. Microbiol. 45 2761–2764
Kellenberger E 2001 Exploring the unknown. EMBO Rep (Vol. 2) (Oxford, UK: Wiley Online Library)
Li W and Godzik A 2006 Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22 1658–1659
Lok C 2015 Mining the microbial dark matter. Nature 522 270–273
Mahé F, Rognes T, Quince C, de Vargas C and Dunthorn M 2014 Swarm: robust and fast clustering method for amplicon-based studies. PeerJ 2 e593
McDonald D, Price MN, Goodrich J, et al. 2012 An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6 610–618
Mercier C, Boyer F, Bonin A and Coissac E 2013 SUMATRA and SUMACLUST: fast and exact comparison and clustering of sequences. Abstr SeqBio 25-26th Nov 2013 27
Metzker ML 2009 Sequencing technologies – the next generation. Nat. Rev. Genet. 11 31
Oakley BB, Fiedler TL, Marrazzo JM and Fredricks DN 2008 Diversity of human vaginal bacterial communities and associations with clinically defined bacterial vaginosis. Appl. Environ. Microbiol. 74 4898–4909
Ounit R, Wanamaker S, Close TJ and Lonardi S 2015 CLARK: fast and accurate classification of metagenomic and genomic sequences using discriminative k-mers. BMC Genomics 16 1–13
Park S, Choi H, Lee B, Chun J, Won J and Yoon S 2018 hc-OTU: A fast and accurate method for clustering operational taxonomic units based on homopolymer compaction. IEEE/ACM Trans. Comput. Biol. Bioinforma 15 441–451
Prabhu P and Duraiswamy K 2013 An efficient visual analysis method for cluster tendency evaluation, data partitioning and internal cluster validation. Comput. Informatics 32 1013–1037
Rognes T, Flouri T, Nichols B, Quince C and Mahé F 2016 VSEARCH: a versatile open source tool for metagenomics. PeerJ 4 e2584
Russell DJ, Way SF, Benson AK and Sayood K 2010 A grammar-based distance metric enables fast and accurate clustering of large sets of 16S sequences. BMC Bioinformatics 11 601
Schloss PD 2010 The effects of alignment quality, distance calculation method, sequence filtering, and region on the analysis of 16S rRNA gene-based studies. PLoS Comput. Biol. 6 19
Schloss PD and Handelsman J 2005 Introducing DOTUR, a computer program for defining opera: onal taxonomic units and Es:ma:ng species richness. Appl. Environ. Microbiol. 71 1501–1506
Schloss PD, Westcott SL, Ryabin T, et al. 2009 Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75 7537–7541
Scholz MB, Lo CC and Chain PSG 2012 Next generation sequencing and bioinformatic bottlenecks: The current state of metagenomic data analysis. Curr. Opin. Biotechnol. 23 9–15
Shokralla S, Spall JL, Gibson JF and Hajibabaei M 2012 Next‐generation sequencing technologies for environmental DNA research. Mol. Ecol. 21 1794–1805
Sun Y, Cai Y, Huse SM, Knight R, Farmerie WG, Wang X and Mai V 2012 A large-scale benchmark study of existing algorithms for taxonomy-independent microbial community analysis. Brief Bioinform. 13 107–121
Sun Y, Cai Y, Liu L, Yu F, Farrell ML, McKendree W and Farmerie W 2009 ESPRIT: estimating species richness using large collections of 16S rRNA shotgun sequences (supplementary data). Nucleic Acids Res. 39 1–18
Turnbaugh PJ, Hamady M, Yatsunenko T, et al. 2009 A core gut microbiom in obese and lean twins. Nature 457 480–484
Van Rijsbergen CJ 1979 Information retrieval (2nd ed.) (Butterworth-Heinemann Newton, MA, USA)
Westcott SL and Schloss PD 2015 De novo clustering methods outperform reference-based methods for assigning 16S rRNA gene sequences to operational taxonomic units. PeerJ 3 e1487
William H.Press, Saul A. Teukoisky, William T. Vetterling BPF 2007 NUMERICAL RECIPES The Art of Scientific Computing (Cambridge University Press)
Woese CR 1987 Bacterial evolution. Microbiol. Rev. 51 221–71
Wood DE and Salzberg SL 2014 Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15 R46
Zou Q, Lin G, Jiang X, Liu X and Zeng X 2018 Sequence clustering in bioinformatics: an empirical study. Brief. Bioinformatics bby090, https://doi.org/10.1093/bib/bby090
Acknowledgements
The authors are grateful to the Management and Principal of K.S.Rangasamy College of Technology, Tiruchengode, Namakkal, Tamil Nadu, India, and DST FIST (Fund for Infrastructure for Science and Technology) FIST No: 368 (SR/FST/College – 235/2014)] and DBT-STAR College Scheme (BT/HRD/11/09/2018) for proving laboratory support in the Institution. Also, we would like to thank Prof. M. Umaiarasi for proof-reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by BJ Rao.
Corresponding editor: BJ Rao
Rights and permissions
About this article
Cite this article
Bhat, A.H., Prabhu, P. & Balakrishnan, K. A critical analysis of state-of-the-art metagenomics OTU clustering algorithms. J Biosci 44, 148 (2019). https://doi.org/10.1007/s12038-019-9964-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12038-019-9964-5