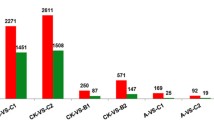
Abstract
Cold acclimation is a complex transcriptionally controlled process regulated by many different genes and genic-interactions in plants. The northward spreading of woody species is mainly limited by winter harshness. To increase our knowledge about the biological processes underlying cold acclimation, plants evolved in warmer climates can serve as models. In this work, a Suppression Subtractive Hybridization approach using PCR-select was used to isolate Italian cypress (Cupressus sempervirens L.) transcript sequences putatively expressed under low temperature stress. After assessing the reliability of the subtractive step, a total of 388 clones were selected and sequenced. Following sequence assembly and removal of the redundant cDNAs, 156 unique transcripts were identified and annotated in order to assign them a putative functional class. Most of the identified transcripts were functionally classified pertaining to stress in cellular and chloroplast membranes, which are previously known to be severely damaged by cold treatment. Among the identified functional gene families, the extensively represented ones were dehydrins, early light-inducible proteins, senescence-associated genes and oleosins. The last three gene families were further selected for phylogenetic analysis, with the corresponding protein sequences across the complete genomes of the model plants Populus trichocarpa, Vitis vinifera, Physcomitrella patens, and Arabidopsis thaliana. The relationship with the ortholog sequences coming from these species and their further implications are discussed.





Similar content being viewed by others
References
Theocharis, A., Clement, C., & Barka, E. A. (2012). Physiological and molecular changes in plants grown at low temperatures. Planta, 235, 1091–1105.
Hincha, D. K. (2002). Cryoprotectin: A plant lipid-transfer protein homologue that stabilizes membranes during freezing. Philosophical Transactions of the Royal Society of London. Series B, 357, 909–915.
Yaish, M. W. F., Doxey, A. C., McConkey, B. J., Moffatt, B. A., & Griffith, M. (2006). Cold-active winter rye glucanases with ice-binding capacity. Plant Physiology, 141, 1459–1472.
Welling, A., & Palva, E. T. (2006). Molecular control of cold acclimation in trees. Physiol Plantarum, 127, 167–181.
Fernandez, O., Theocharis, A., Bordiec, S., Feil, R., Jacquens, L., Clement, C., et al. (2012). Acclimates grapevine to cold by modulating carbohydrate metabolism. Molecular Plant-Microbe Interactions, 25, 496–504.
Eriksson, S. K., Kutzer, M., Procek, J., Grobner, G., & Harryson, P. (2011). Tunable membrane binding of the intrinsically disordered dehydrin Lti30, a cold-induced plant stress protein. Plant Cell, 23, 2391–2404.
Bies-Etheve, N., Gaubier-Comella, P., Debures, A., Lasserre, E., Jobet, E., Raynal, M., et al. (2008). Inventory, evolution and expression profiling diversity of the LEA (late embryogenesis abundant) protein gene family in Arabidopsis thaliana. Plant Molecular Biology, 67, 107–124.
Heddad, M., Noren, H., Reiser, V., Dunaeva, M., Andersson, B., & Adamska, I. (2006). Differential expression and localization of early light-induced proteins in Arabidopsis. Plant Physiology, 142, 75–87.
Degenkolbe, T., Giavalisco, P., Zuther, E., Seiwert, B., Hincha, D. K., & Willmitzer, L. (2012). Differential remodeling of the lipidome during cold acclimation in natural accessions of Arabidopsis thaliana. The Plant Journal, 72, 972–982.
Mao, D., & Chen, C. (2012). Colinearity and similar expression pattern of rice DREB1 s reveal their functional conservation in the cold-responsive pathway. PLoS ONE, 7, e47275. doi:10.1371/journal.pone.0047275.
Checker, V. G., Chhibbar, A. K., & Khurana, P. (2012). Stress-inducible expression of barley Hva1 gene in transgenic mulberry displays enhanced tolerance against drought, salinity and cold stress. Transgenic Research, 21, 939–957.
Perdiguero, P., Barbero, M. C., Cervera, M. T., Soto, A., & Collada, C. (2012). Novel conserved segments are associated with differential expression patterns for Pinaceae dehydrins. Planta, 236, 1863–1874.
Dauwe, R., Holliday, J. A., Aitken, S. N., & Mansfield, S. D. (2012). Metabolic dynamics during autumn cold acclimation within and among populations of Sitka spruce (Picea sitchensis). New Phytologist, 194, 192–205.
D’Angeli, S., Falasca, G., Matteucci, M., & Altamura, M. M. (2013). Cold perception and gene expression differ in Olea europaea seed coat and embryo during drupe cold acclimation. New Phytologist, 197, 123–138.
Larcher, W. (2001). Ökophysiologie der Pflanzen: (S. E. Ulmer) ed (p. 302). Stuttgart: Eugen Ulmer.
Bagnoli, F., Vendramin, G. G., Buonamici, A., Doulis, A. G., Gonzalez-Martinez, S. C., La Porta, N., et al. (2009). Is Cupressus sempervirens native in Italy? An answer from genetic and palaeobotanical data. Molecular Ecology, 18, 2276–2286.
Baldi, P., Muthuchelian, K., & La Porta, N. (2012). Leaf plasticity to light intensity in Italian cypress (Cupressus sempervirens L.): Adaptability of a Mediterranean conifer cultivated in the Alps. Journal of Photochemistry and Photobiology B: Biology, 117, 61–69.
Zorer, R., Eccel, E., Tognetti, R., & La Porta, N. (2014). A Mediterranean conifer under vegetation shift: Seasonal changes of photochemical activity in Cupressus sempervirens (L.) and evidence of correlation with temperature models. Italian Journal of Agrometeorology, 1, 29–40.
La Porta, N., Bertamini, M., Nedunchezhian, N., Raddi, P., & Muthuchelian, K. (2005). Photoinhibition of photosynthesis in needles of two cypress (Cupressus sempervirens L.) clones. Tree Physiology, 25, 1033–1039.
Weger, H. G., Silim, S. N., & Guy, R. D. (1993). Photosynthetic acclimation to low temperature by western red cedar seedlings. Plant, Cell and Environment, 16, 711–717.
La Porta, N., Bertamini, M., Nedunchezhian, N., & Muthuchelian, K. (2006). Photosynthetic changes that occur during aging of cypress (Cupressus sempervirens L.) needles. Photosynthetica, 44, 555–560.
Muthuchelian, K., La Porta, N., Bertamini, M., & Nedunchezhian, N. (2005). Cypress canker induced inhibition of photosynthesis in field grown cypress (Cupressus sempervirens L.) needles. Physiological and Molecular Plant Pathology, 67, 33–39.
Muthuchelian, K., Bertamini, M., La Porta, N., & Nedunchezhian, N. (2005). Photoinhibition and recovery of photosynthesis in canker-susceptible and resistant needles of cypress (Cupressus sempervirens L.). J. Phytopath., 153, 337–343.
Zocca, A., Zanini, C., Aimi, A., Frigimelica, G., La Porta, N., & Battisti, A. (2008). Spread of a plant pathogen and associated insect vectors at the northern range margin of cypress (Cupressus sempervirens). Acta Oecologica, 33, 307–313.
Oshlack, A., Robinson, M. D., & Young, M. D. (2010). From RNA-seq reads to differential expression results. Genome Biology, 11, 220.
Gu, R. S., Fonseca, S., Puskas, L. G., Hackler, L., Zvara, A., Dudits, D., & Pais, M. S. (2004). Transcript identification and profiling during salt stress and recovery of Populus euphratica. Tree Physiology, 24, 265–276.
Yakovlev, I. A., Fossdal, C. G., Johnsen, O., Junttila, O., & Skroppa, T. (2006). Analysis of gene expression during bud burst initiation in Norway spruce via ESTs from subtracted cDNA libraries. Tree Genetics and Genomes, 2, 39–52.
Pedron, L., Baldi, P., Hietala, A. M., & La Porta, N. (2009). Genotype-specific regulation of cold-responsive genes in cypress (Cupressus sempervirens L.). Gene, 437, 45–53.
Xu, L., Yuan, Y., Zhang, L., Wan, L., Zheng, Y., Zhou, P., & Li, D. (2011). Identification and characterization of differential gene expression in the mesocarp and kernel of oil palm nuts using suppression subtractive hybridization. Tree Genetics and Genomes, 7, 999–1010.
La Porta, N., Battisti, A., & Raddi, P. (2005). Ecological assessment and sustainable management of cypress under climate change conditions in the Italian Alps. International Forest Review, 7(5), 82.
Moser, C., Gatto, P., Moser, M., Pindo, M., & Velasco, R. (2004). Isolation of functional RNA from small amounts of different grape and apple tissues. Molecular Biotechnology, 26, 95–99.
Simon, N., Campbell, L., Ornolfsdottir, E., Groben, R., Guillou, L., Lange, M., & Medlin, L. K. (2000). Oligonucleotide probes for the identification of three algal groups by dot blot and fluorescent whole-cell hybridization. Journal of Eukaryotic Microbiology, 47, 76–84.
Edgar, R. C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32, 1792–1797.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., & Kumar, S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28, 2731–2739.
Huang, X., & Madan, A. (1999). CAP3: A DNA sequence assembly program. Genome Research, 9, 868–877.
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet, 25, 25–29.
Schiex, T., Gouzy, J., Moisan, A., & De Oliveira, Y. (2003). FrameD: A flexible program for quality check and gene prediction in prokaryotic genomes and noisy matured eukaryotic sequences. Nucleic Acids Research, 31, 3738–3741.
Simkin, A. J., Qian, T., Caillet, V., Michoux, F., Ben Amor, M., Lin, C., et al. (2006). Oleosin gene family of Coffea canephora: Quantitative expression analysis of five oleosin genes in developing and germinating coffee grain. Journal of Plant Physiology, 163, 691–708.
Peng, Y., Lin, W., Wei, H., Krebs, S. L., & Arora, R. (2008). Phylogenetic analysis and seasonal cold acclimation-associated expression of early light-induced protein genes of Rhododendron catawbiense. Physiologia Plantarum, 132, 44–52.
Palmé, A. E., Pyhäjärvi, T., Wachowiak, W., & Savolainen, O. (2009). Selection on nuclear genes in a Pinus phylogeny. Molecular Biology and Evolution, 26, 893–905.
Thomashow, M. F. (1999). Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annual Review of Plant Physiology and Plant Molecular Biology, 50, 571–599.
Wang, Y. Q., Shen, J. K., Berglund, T., Ohlsson, A. B., Tang, X. F., Zhou, Z. K., et al. (2010). Analysis of expressed sequence tags from Ginkgo mature foliage in China. Tree Genetics and Genomes, 6, 357–365.
Ujino-Ihara, T., Kanamori, H., Yamane, H., Taguchi, Y., Namiki, N., Mukai, Y., et al. (2005). Comparative analysis of expressed sequence tags of conifers and angiosperms reveals sequences specifically conserved in conifers. Plant Molecular Biology, 59, 895–907.
Mishra, S., Srivastava, S., & Nautiyal, C. S. (2014). Differential gene expression profile in Pseudomonas putida NBRIC19-treated wheat (Triticum aestivum) plants subjected to biotic stress of Parthenium hysterophorus. Molecular Biology Reports, 41, 1385–1399.
Fan, Q. J., Yan, F. X., Qiao, G., Zhang, B. X., & Wen, X. P. (2014). Identification of differentially-expressed genes potentially implicated in drought response in pitaya (Hylocereus undatus) by suppression subtractive hybridization and cDNA microarray analysis. Gene, 533, 322–331.
Kaplan, F., Kopka, J., Sung, D. Y., Zhao, W., Popp, M., Porat, R., & Guy, C. L. (2007). Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. The Plant Journal, 50, 967–981.
Purdy, S. J., Bussell, J. D., Nunn, C. P., & Smith, S. M. (2013). Leaves of the Arabidopsis maltose exporter1 mutant exhibit a metabolic profile with features of cold acclimation in the warm. PLoS One, 8, e79412. doi:10.1371/journal.pone.0079412.
Lee, J. H., Yu, D. J., Kim, S. J., Choi, D., & Lee, H. J. (2012). Intraspecies differences in cold hardiness, carbohydrate content and beta-amylase gene expression of Vaccinium corymbosum during cold acclimation and deacclimation. Tree Physiology, 32, 1533–1540.
Ito, A., Sugiura, T., Sakamoto, D., & Moriguchi, T. (2013). Effects of dormancy progression and low-temperature response on changes in the sorbitol concentration in xylem sap of Japanese pear during winter season. Tree Physiology, 33, 398–408.
Secchi, F., & Zwieniecki, M. A. (2012). Analysis of xylem sap from functional (non-embolized) and nonfunctional (embolized) vessels of Populus nigra: Chemistry of refilling. Plant Physiology, 160, 955–964.
Riikonen, J., Kontunen-Soppela, S., Vapaavuori, E., Tervahauta, A., Tuomainen, M., & Oksanen, E. (2013). Carbohydrate concentrations and freezing stress resistance of silver birch buds grown under elevated temperature and ozone. Tree Physiology, 33, 311–319.
Boneh, U., Biton, I., Schwartz, A., & Ben-Ari, G. (2012). Characterization of the ABA signal transduction pathway in Vitis vinifera. Plant Science, 187, 89–96.
Mittler, R. (2006). Abiotic stress, the field environment and stress combination. Trends in Plant Science, 11, 15–19.
Mao, X., Chen, S., Li, A., Zhai, C., & Jing, R. (2014). Novel NAC transcription factor TaNAC67 confers enhanced multi-abiotic stress tolerances in Arabidopsis. PLoS One, 9, e84359. doi:10.1371/journal.pone.0084359.
Baldi, P., Pedron, L., Hietala, A. M., & La Porta, N. (2011). Cold tolerance in cypress (Cupressus sempervirens L.): A physiological and molecular study. Tree Genetics and Genomes, 7, 79–90.
Uemura, M., Tominaga, Y., Nakagawara, C., Shigematsu, S., Minami, A., & Kawamura, Y. (2006). Responses of the plasma membrane to low temperatures. Physiologia Plantarum, 126, 81–89.
Kazemi Shahandashti, S. S., Maali Amiri, R., Zeinali, H., & Ramezanpour, S. S. (2013). Change in membrane fatty acid compositions and cold-induced responses in chickpea. Molecular Biology Reports, 40, 893–903.
Abdrakhamanova, A., Wang, Q. Y., Khokhlova, L., & Nick, P. (2003). Is microtubule disassembly a trigger for cold acclimation? Plant and Cell Physiology, 44, 676–686.
Kosova, K., Vitamvas, P., & Prasil, I. T. (2007). The role of dehydrins in plant response to cold. Biologia Plantarum, 51, 601–617.
Sandve, S. R., Kosmala, A., Rudi, H., Fjellheim, S., Rapacz, M., Yamada, T., & Rognli, O. A. (2011). Molecular mechanisms underlying frost tolerance in perennial grasses adapted to cold climates. Plant Science, 180, 69–77.
La Porta, N., Bertamini, M., Nedunchezhian, N., & Muthuchelian, K. (2004). High irradiance induced changes on photosystem 2 in young and mature needles of cypress (Cupressus sempervirens L.). Photosynthetica, 42, 263–271.
Bascunan-Godoy, L., Sanhueza, C., Cuba, M., Zuniga, G. E., Corcuera, L. J., & Bravo, L. A. (2012). Cold-acclimation limits low temperature induced photoinhibition by promoting a higher photochemical quantum yield and a more effective PSII restoration in darkness in the Antarctic rather than the Andean ecotype of Colobanthus quitensis Kunt Bartl (Cariophyllaceae). BMC Plant Biology, 12, 114.
Nylander, M., Svensson, J., Palva, E. T., & Welin, B. V. (2001). Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Molecular Biology, 45, 263–279.
Rorat, T. (2006). Plant dehydrins-tissue location, structure and function. Cellular & Molecular Biology Letters, 11, 536–556.
Yang, Y., He, M., Zhu, Z., Li, S., Xu, Y., Zhang, C., et al. (2012). Identification of the dehydrin gene family from grapevine species and analysis of their responsiveness of various forms of abiotic and biotic stress. BMC Plant Biology, 54, 743–753.
Hanin, M., Brini, F., Ebel, E., Toda, Y., Takeda, S., & Masmoudi, K. (2011). Plant dehydrins and stress tolerance: Versatile proteins for complex mechanisms. Plant Signaling & Behavior, 6, 1503–1509.
Adamska, I., & Kloppstech, K. (1991). Evidence for an association of the early light-inducible protein (ELIP) of pea with photosystem II. Plant Molecular Biology, 16, 209–223.
Hutin, C., Nussaume, L., Moise, N., Moya, I., Kloppstech, K., & Havaux, M. (2003). Early light-induced proteins protect Arabidopsis from photooxidative stress. Proceedings of the National Academy of Sciences of the United States of America, 100, 4921–4926.
Zarter, C. R., Adams, W. W, 3rd, Ebbert, V., Adamska, I., Jansson, S., & Demmig-Adams, B. (2006). Winter acclimation of PsbS and related proteins in the evergreen Arctostaphylos uva-ursi as influenced by altitude and light environment. Plant, Cell and Environment, 29, 869–878.
Rossini, S., Casazza, A. P., Engelmann, E. C., Havaux, M., Jennings, R. C., & Soave, C. (2006). Suppression of both ELIP1 and ELIP2 in Arabidopsis does not affect tolerance to photoinhibition and photooxidative stress. Plant Physiology, 141, 1264–1273.
Zhou, C. G. S. (2009). Senescence. In D. M. Pua EC (Ed.), Plant developmental biology: Biotechnological perspectives (pp. 151–169). Berlin: Springer.
Guo, Y., & Gan, S. S. (2012). Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence-promoting hormonal, pathological and environmental stress treatments. Plant, Cell and Environment, 35, 644–655.
Zhao, L., Zhang, H., Zhang, B., Bai, X., & Zhou, C. (2012). Physiological and molecular changes of detached wheat leaves in responding to various treatments. Journal of Integrative Plant Biology, 54, 567–576.
Lim, P. O., Kim, H. J., & Nam, H. G. (2007). Leaf senescence. Annual Review of Plant Biology, 58, 115–136.
Shimada, T. L., Shimada, T., Takahashi, H., Fukao, Y., & Hara-Nishimura, I. (2008). A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. The Plant Journal, 55, 798–809.
Tanaka, H., Osakabe, Y., Katsura, S., Mizuno, S., Maruyama, K., Kusakabe, K., et al. (2012). Abiotic stress-inducible receptor-like kinases negatively control ABA signaling in Arabidopsis. The Plant Journal, 70, 599–613.
Xue-Xuan, X., Hong-Bo, S., Yuan-Yuan, M., Gang, X., Jun-Na, S., Dong-Gang, G., & Cheng-Jiang, R. (2010). Biotechnological implications from abscisic acid (ABA) roles in cold stress and leaf senescence as an important signal for improving plant sustainable survival under abiotic-stressed conditions. Critical Reviews in Biotechnology, 30, 222–230.
Huang, G. T., Ma, S. L., Bai, L. P., Zhang, L., Ma, H., Jia, P., et al. (2012). Signal transduction during cold, salt, and drought stresses in plants. Molecular Biology Reports, 39, 969–987.
Tzen, J. T., Lai, Y. K., Chan, K. L., & Huang, A. H. (1990). Oleosin isoforms of high and low molecular weights are present in the oil bodies of diverse seed species. Plant Physiology, 94, 1282–1289.
Kim, H. U., Hsieh, K., Ratnayake, C., & Huang, A. H. (2002). A novel group of oleosins is present inside the pollen of Arabidopsis. Journal of Biological Chemistry, 277, 22677–22684.
Acknowledgments
The authors wish to thank Dr. Paolo Raddi, Mr. Vincenzo Di Lonardo, and Dr. Roberto Danti (IPP-CNR, Florence, Italy) who kindly supplied the selected cypress genotypes used in the experiments. We gratefully thank also Mr. Federico Piccirillo (Montemagno Resource Inc.), Mr. Emanuel Endrizzi and Dr. Luca Pedron (Fondazione Edmund Mach) for the technical assistance in the laboratory and field and Mrs. Tessa Say for the useful English corrections. The authors acknowledge the two anonymous reviewers and the editor for the appreciated constructive comments and suggestions which have helped to improve the manuscript. This work is part of the project ECOCYPRE (Ecological assessment and sustainable managements of cypress in the landscape of Trentino) that was funded to Dr. Nicola La Porta by the Provincia autonoma di Trento through Grant No. 437.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
La Porta, N., Sablok, G., Emilliani, G. et al. Identification of Low Temperature Stress Regulated Transcript Sequences and Gene Families in Italian Cypress. Mol Biotechnol 57, 407–418 (2015). https://doi.org/10.1007/s12033-014-9833-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-014-9833-2