Abstract
Background/Objective
Iron can be detrimental to most tissues both in excess and in deficiency. The brain in particular is highly susceptible to the consequences of excessive iron, especially during blood brain barrier disruption after injury. Preliminary evidence suggests that iron homeostasis is important during recovery after neurologic injury; therefore, the exploration of genetic variability in genes involved in iron homeostasis is an important area of patient outcomes research. The purpose of this study was to examine the relationship between tagging single nucleotide polymorphisms (SNPs) in candidate genes related to iron homeostasis and acute and long-term patient outcomes after aneurysmal subarachnoid hemorrhage (aSAH).
Methods
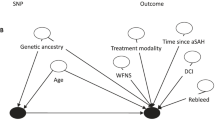
This study was a longitudinal, observational, candidate gene association study of participants with aSAH that used a two-tier design including tier 1 (discovery, n = 197) and tier 2 (replication, n = 277). Participants were followed during the acute outcome phase for development of cerebral vasospasm and delayed cerebral ischemia (DCI) and during the long-term outcome phase for death and gross functional outcome using the Glasgow Outcome Scale (GOS; poor = 1–3). Genetic association analyses were performed using a logistic regression model adjusted for age, sex, and Fisher grade. Approximate Bayes factors (ABF) and Bayesian false discovery probabilities (BFDP) were used to prioritize and interpret results.
Results
In tier 1, 235 tagging SNPs in 28 candidate genes were available for analysis and 26 associations (20 unique SNPs in 12 genes) were nominated for replication in tier 2. In tier 2, we observed an increase in evidence of association for three associations in the ceruloplasmin (CP) and cubilin (CUBN) genes. We observed an association of rs17838831 (CP) with GOS at 3 months (tier 2 results, odds ratio [OR] = 2.10, 95% confidence interval [CI] = 1.14–3.86, p = 0.018, ABF = 0.52, and BFDP = 70.8%) and GOS at 12 months (tier 2 results, OR = 1.86, 95% CI 0.98–3.52, p = 0.058, ABF = 0.72, and BFDP = 77.3%) as well as rs10904850 (CUBN) with DCI (tier 2 results, OR = 0.70, 95% CI 0.48–1.02, p = 0.064, ABF = 0.59, and BFDP = 71.8%).
Conclusions
Among the genes examined, our findings support a role for CP and CUBN in patient outcomes after aSAH. In an effort to translate these findings into clinical utility and improve outcomes after aSAH, additional research is needed to examine the functional roles of these genes after aSAH.

Similar content being viewed by others
References
Boling B, Groves TR. Management of subarachnoid hemorrhage. Crit Care Nurse. 2019;39(5):58–67.
Zacharia BE, Hickman ZL, Grobelny BT, et al. Epidemiology of aneurysmal subarachnoid hemorrhage. Neurosurg Clin N Am. 2010;21(2):221–33.
van Gijn J, Kerr RS, Rinkel GJE. Subarachnoid haemorrhage. Lancet. 2007;369(9558):306–18.
Teo M, Teo M, Turner C, et al. What factors determine treatment outcome in aneurysmal subarachnoid hemorrhage in the modern era? A Post Hoc STASH analysis. World Neurosurg. 2017;105:270–81.
Tan G, Liu L, He Z, Sun J, Xing W, Sun X. Role of hepcidin and its downstream proteins in early brain injury after experimental subarachnoid hemorrhage in rats. Mol Cell Biochem. 2016;418(1):31–8.
Gomes JA, Selim M, Cotleur A, et al. Brain iron metabolism and brain injury following subarachnoid hemorrhage: iCeFISH-pilot (CSF iron in SAH). Neurocrit Care. 2014;21(2):285–93.
Heinsberg LW, Arockiaraj AI, Crago EA, et al. Genetic variability and trajectories of DNA methylation may support a role for HAMP in patient outcomes after aneurysmal subarachnoid hemorrhage. Neurocrit Care 2019;32(2):550–63.
Wagner KR, Sharp FR, Ardizzone TD, Lu A, Clark JF. Heme and iron metabolism: role in cerebral hemorrhage. J Cereb Blood Flow Metab. 2003;23(6):629–52.
Bishop GM, Robinson SR. Quantitative analysis of cell death and ferritin expression in response to cortical iron: implications for hypoxia-ischemia and stroke. Brain Res. 2001;907(1–2):175–87.
Garton T, Keep RF, Hua Y, Xi G. Brain iron overload following intracranial haemorrhage. Stroke Vasc Neurol. 2016;1(4):172–84.
Kim H, Crago E, Kim M, et al. Cerebral vasospasm after sub-arachnoid hemorrhage as a clinical predictor and phenotype for genetic association study. Int J Stroke. 2013;8(8):620–5.
Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75.
Purcell S. PLINK. Available from: https://www.cog-genomics.org/plink2. Accessed 20 Mar 2020.
Team RC. R: a language and environment for statistical computing. 2018; Available from: https://www.r-project.org/. Accessed 22 Jan 2018.
Meyer H. plinkQC: genotype quality control with “PLINK.” 2019.
Marees AT, de Kluiver H, Stringer S, et al. A tutorial on conducting genome-wide association studies: quality control and statistical analysis. Int J Methods Psychiatr Res. 2018;27(2):e1608.
Karolchik D, Hinrichs A, Furey T, et al. The UCSC table browser data retrieval tool. Nucleic Acids Res. 2004;32:493–6.
Machiela M, Chanock S. LDlink a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31:3555–7.
Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215.
Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the sequenom massARRAY iPLEX platform. Curr Protoc Hum Genet. 2009;60(suppl 60):2121–21218. https://doi.org/10.1002/0471142905.hg0212s60.
Jennett B, Bond M. Assessment of outcome after severe brain damage. Lancet (London, England). 1975;1(7905):480–4.
Wakefield J. A Bayesian measure of the probability of false discovery in molecular genetic epidemiology studies. Am J Hum Genet. 2007;81:208–27.
Wakefield J. Bayes factors for genome-wide association studies: comparison with P values. Genet Epidemiol. 2008;33(1):79–86.
Qian Y, Yin C, Chen Y, et al. Estrogen contributes to regulating iron metabolism through governing ferroportin signaling via an estrogen response element. Cell Signal. 2015;27(5):934–42.
Patel BN, Dunn RJ, Jeong SY, Zhu Q, Julien JP, David S. Ceruloplasmin regulates iron levels in the CNS and prevents free radical injury. J Neurosci. 2002;22(15):6578–86.
Besold AN, Culbertson EM, Culotta VC. The Yin and Yang of copper during infection. J Biol Inorg Chem. 2016;21(2):137–44.
Cherukuri S, Potla R, Sarkar J, Nurko S, Harris ZL, Fox PL. Unexpected role of ceruloplasmin in intestinal iron absorption. Cell Metab. 2005;2(5):309–19.
Bickford JS, Ali NF, Nick JA, et al. Endothelin-1-mediated vasoconstriction alters cerebral gene expression in iron homeostasis and eicosanoid metabolism. Brain Res. 2014;1588:25–36.
Adamsson Eryd S, Sjögren M, Smith JG, et al. Ceruloplasmin and atrial fibrillation: evidence of causality from a population-based Mendelian randomization study. J Intern Med. 2014;275(2):164–71. https://doi.org/10.1111/joim.12144.
Verroust PJ, Christensen EI. Megalin and cubilin–the story of two multipurpose receptors unfolds. Nephrol Dial Transpl. 2002;17(11):1867–71.
Böger CA, Chen MH, Tin A, et al. CUBN is a gene locus for albuminuria. J Am Soc Nephrol. 2011;22(3):555–70.
McLaren CE, McLachlan S, Garner CP, et al. Associations between single nucleotide polymorphisms in iron-related genes and iron status in multiethnic populations. PLoS ONE. 2012;7(6):e338339.
Zacharia BE, Ducruet AF, Hickman ZL, et al. Renal dysfunction as an independent predictor of outcome after aneurysmal subarachnoid hemorrhage: a single-center cohort study. Stroke. 2009;40(7):2375–81.
Hamdan A, Barnes J, Mitchell P. Subarachnoid hemorrhage and the female sex: Analysis of risk factors, aneurysm characteristics, and outcomes. J Neurosurg. 2014;121(6):1367–73.
Morton MJ, Hostettler IC, Kazmi N, et al. Haptoglobin genotype and outcome after aneurysmal subarachnoid haemorrhage. J Neurol Neurosurg Psychiatry. 2019;91(3):305–13.
Hugelshofer M, Buzzi RM, Schaer CA, et al. Haptoglobin administration into the subarachnoid space prevents hemoglobin-induced cerebral vasospasm. J Clin Invest. 2019;129(12):5219–35.
Acknowledgements
We would like to acknowledge Sandra Deslouches for her expertise and work in the laboratory, Tiffany Wang for her help formatting the Supplemental Tables associated with this publication, and the anonymous reviewers who took the time to critically evaluate this paper as their feedback improved the clarity and quality of this work.
Funding
Research reported in this publication was supported by the National Institute of Nursing Research of the National Institutes of Health under Award Nos. F31NR017311, R01NR004339, R01NR013610, and T32NR009759 with additional support from the Nightingale Awards of Pennsylvania, Center for Jonas Nursing and Veterans Healthcare, Jayne F. Wiggins Memorial Award, and Sigma Theta Tau—Eta Chapter. The content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health or supporting foundations.
Author information
Authors and Affiliations
Contributions
All authors meet authorship criteria, have read and approved the submitted manuscript, and certify that they have participated sufficiently in the work to take responsibility for the content including the concept, design, analysis, writing, or revision. Lacey W. Heinsberg contributed to the study conception and design, acquisition, analysis, and interpretation of data, and drafted, critically revised, and gave final approval for the manuscript. Sheila A. Alexander contributed to the study design, interpretation of data, and critically revised and gave final approval for the manuscript. Elizabeth A. Crago contributed to the acquisition and interpretation of data and critically revised and gave final approval for the manuscript. Ryan L. Minster contributed to the analysis and interpretation of data and critically revised and gave final approval for the manuscript. Samuel M. Poloyac contributed to acquisition and interpretation of data and critically revised and gave final approval for the manuscript. Daniel E. Weeks contributed to the study design, analysis and interpretation of data, and critically revised and gave final approval for the manuscript. Yvette P. Conley contributed to the study conception and design, acquisition and interpretation of data, and critically revised and gave final approval for the manuscript. All authors agree to be accountable for all aspects of the work in ensuring that questions relating to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Corresponding author
Ethics declarations
Conflict of interest
LW Heinsberg reports Grants from the National Institutes of Health, University of Pittsburgh Jayne F. Wiggins Memorial Scholarship, Eta Chapter, Sigma Theta Tau, Inc., Jonas Foundation, and the Nightingale Awards of Pennsylvania during the conduct of this study. YP Conley, DE Weeks, and EA Crago reports Grants from the National Institutes of Health. SA Alexander, RL Minster, and SM Poloyac report nothing to disclose.
Ethical Conduct of Research
Informed consent was obtained from all study participants. Institutional Review Board approval at the University of Pittsburgh is in place (IRB approval number STUDY19100368) and we have adhered to ethical considerations in the protection of all human subjects involved.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Heinsberg, L.W., Alexander, S.A., Crago, E.A. et al. Genetic Variability in the Iron Homeostasis Pathway and Patient Outcomes After Aneurysmal Subarachnoid Hemorrhage. Neurocrit Care 33, 749–758 (2020). https://doi.org/10.1007/s12028-020-00961-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12028-020-00961-z