Abstract
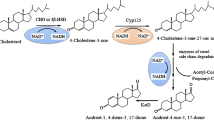
In biotransformation of phytosterol to 4-androstene-3,17-dione (AD) by Mycobacterium, the steroidal alcohol (such as 22-hydroxy-23,24-bisnorchol-4-ene-3-one, HBC) was a main byproduct. To weaken the accumulation of this byproduct in sterol biotransformation, ammonium was substituted by nitrate as nitrogen resource. The nitrate was utilized by Mycobacterium and led to metabolic flux shift towards AD production. The ratio of AD/HBC increased maximally from 2.1 to 5.5 and AD production increased correspondently. In the meanwhile, the nitrate metabolism resulted in the decreased intracellular redox level (NADH/NAD+) maximally by 59.5% and a slight descent tendency with the increase of the nitrate concentrations. It indicated that the nitrate utilization effectively decreased the steroidal alcohol production by regulating intracellular redox level in sterol biotransformation. These results gave an insight into the mechanism of the steroidal alcohol formation in sterol biodegradation and provided a simple strategy to regulate the metabolic distribution.



Similar content being viewed by others
References
Donova, M. V., & Egorova, O. V. (2012). Microbial steroid transformations: current state and prospects. Applied Microbiology and Biotechnology, 94(6), 1423–1447.
Bhatti, H. N., & Khera, R. A. (2012). Biological transformations of steroidal compounds: a review. Steroids, 77(12), 1267–1290.
Wei, W., Fan, S. F., Wang, F. Q., & Wei, D. Z. (2010). A new steroid-transforming strain of Mycobacterium neoaurum and cloning of 3-Ketosteroid 9α-Hydroxylase in NwIB-01. Applied Biochemistry and Biotechnology, 162(5), 1446–1456.
Szentirrnai, A. (1990). Microbial physiology of sidechain degradation of sterols. Journal of Industrial Microbiology & Biotechnology, 6, 101–116.
Su, L. Q., Shen, Y. B., Zhang, W. K., Gao, T., Shang, Z. H., & Wang, M. (2017). Cofactor engineering to regulate NAD+/NADH ratio with its application to phytosterols biotransformation. Microbial Cell Factoty, 16, 182.
Mädje, K., Schmölzer, K., Nidetzky, B., & Kratzer, R. (2012). Host cell and expression engineering for development of an E. coli ketoreductase catalyst: enhancement of formate dehydrogenase activity for regeneration of NADH. Microbial Cell Factories, 11(1), 7.
Chen, X. L., Li, S. B., & Liu, L. M. (2014). Engineering redox balance through cofactor systems. Trends in Biotechnology, 32(6), 337–343.
Geertman, J. M., van Dijken, J. P., & Pronk, J. T. (2006). Engineering NADH metabolism in Saccharomyces cerevisiae: formate as an electron donor for glycerol production by anaerobic, glucose-limited chemostat cultures. FEMS Yeast Research, 6(8), 1193–1203.
Sánchez, A. M., Bennett, G. N., & San, K. Y. (2005). Effect of different levels of NADH availability on metabolic fluxes of Escherichia coli chemostat cultures in defined medium. Journal of Bacteriology, 117, 395–405.
Heux, S., Cachon, R., & Dequin, S. (2006). Cofactor engineering in Saccharomyces cerevisiae: expression of a H2O-forming NADH oxidase and impact on redox metabolism. Metabolic Engineering, 8(4), 303–314.
Lu, H. Z., Liu, X. Y., Huang, M. Z., Xi, A. J. Y., Zhuang, Y. P., Zhang, S. L., & Noorman, H. (2015). Integrated isotope-assisted metabolomics and 13C metabolic flux analysis reveals metabolic flux redistribution for high glucoamylase production by Aspergillus niger. Microbial Cell Factoty, 14, 147.
Susana, J., Berríos-Rivera, K. Y., San, G. N., & Bennett, G. N. (2002). The effect of NAPRTase overexpression on the total levels of NAD, the NADH/NAD+ ratio, and the distribution of metabolites in Escherichia coli. Metabolic Engineering, 4(3), 238–247.
Xu, X. W., Gao, X. Q., Feng, J. X., Wang, X. D., & Wei, D. Z. (2015). Influence of temperature on nucleus degradation of 4-androstene-3, 17-dione in phytosterol biotransformation by Mycobacterium sp. Letters in Applied Microbiology, 61(1), 63–68.
Gao, X. Q., Feng, J. X., Hua, Q., Wei, D. Z., & Wang, X. D. (2014). Investigation of factors affecting biotransformation of phytosterols to 9-hydroxyandrost-4-ene-3,17-dione based on the HP-β-CD-resting cells reaction system. Biocatalysis and Biotransformation, 32, 343–347.
San, K. Y., Bennett, G. N., Berrís-Rivera, S. J., Vadali, R. V., Yang, Y. T., Horton, E., Rudolph, F. B., Sariyar, B., & Blackwood, K. (2002). Metabolic engineering through cofactor manipulation and its effects on metabolic flux redistribution in Escherichia coli. Metabolic Engineering, 4, 182–192.
Toya, Y., Nakahigashi, K., Tomita, M., & Shimizu, K. (2012). Metabolic regulation analysis of wild-type and arcA mutant Escherichia coli under nitrate conditions using different levels of omics data. Molecular BioSystems, 8(10), 2593–2604.
De Graef, M. R., Alexeeva, S., Snoep, J. L., & de Mattos, M. J. (1999). The steady-state internal redox state (NADH/NAD) reflects the external redox state and is correlated with catabolic adaptation in Escherichia coli. Journal of Bacteriology, 181(8), 2351–2357.
Leonardo, M. R., Cunningham, P. R., & Clark, D. (1993). Anaerobic regulation of the adhE gene, encoding the fermentative alcohol dehydrogenase of Escherichia coli. Journal of Bacteriology, 175(3), 870–878.
Leonardo, M. R., Dailly, Y., & Clark, D. (1996). Role of NAD in regulating the adhE gene of Escherichia coli. Journal of Bacteriology, 178(20), 6013–6018.
Hansen, H. G., & Henning, U. (1996). Regulation of pyruvate dehydrogenase activity in Escherichia coli K12. Biochimica et Biophysica Acta (BBA)- Enzymology & Biological Oxidation, 122(2), 355–358.
Funding
This work was supported by the National Natural Science Foundation of China (31570079, 21276083).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, X., Chen, R., Wu, Y. et al. Nitrate Metabolism Decreases the Steroidal Alcohol Byproduct Compared with Ammonium in Biotransformation of Phytosterol to Androstenedione by Mycobacterium neoaurum. Appl Biochem Biotechnol 190, 1553–1560 (2020). https://doi.org/10.1007/s12010-019-03175-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-019-03175-y