Abstract
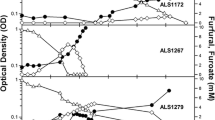
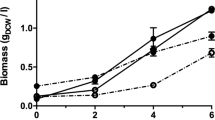
The genes involved in the aerobic bacterial metabolism of furfural and 5-hydroxymethylfurfural (HMF) have been characterized in two species, Pseudomonas putida Fu1 and Cupriavidus basilensis HMF14. A third furan-metabolizing strain, Pseudomonas putida ALS1267, was recently identified that grows robustly on both furfural and HMF as sole carbon sources, with a growth rate of 0.250 h−1 on furfural and 0.311 h−1 on HMF, and we have characterized the genes involved in furfural and HMF metabolism in this bacterium. Unlike C. basilensis HMF14, which contains separate furfural and HMF operons, P. putida ALS1267 contains one contiguous 18.1-kb operon, which harbors all of the furfural- and HMF-metabolizing genes except for one, the hmfH gene that encodes HMF/furfural oxidoreductase and has both HMF acid oxidase and furfural/HMF dehydrogenase activity. The 18.1-kb operon was cloned into P. putida KT2440, which cannot metabolize furans, enabling growth on furfural as the sole carbon source with a growth rate of 0.340 h−1. The clone did not allow P. putida KT2440 to metabolize HMF, most likely due to the lack of the hmfH gene. No hmfH homolog was identified in P. putida ALS1267, suggesting that another gene in the ALS1267 genome provides this function.



Similar content being viewed by others
References
Boyer, L. J., Vega, J. L., Klasson, K. T., Clausen, E. C., & Gaddy, J. L. (1992). The effects of furfural on ethanol production by Saccharomyces cerevisiae in batch culture. Biomass and Bioenergy, 3(1), 41–48.
Delgenes, J. P., Moletta, R., & Navarro, J. M. (1996). Effects of lignocellulose degradation products on ethanol fermentations of glucose and xylose by Saccharomyces cerevisiae, Zymomonas mobilis, Pichia stipitis, and Candida shehatae. Enzyme and Microbial Technology, 19(3), 220–225.
Larsson, S., Palmqvist, E., Hahn-Hägerdal, B., Tengborg, C., Stenberg, K., Zacchi, G., & Nilvebrant, N. O. (1999). The generation of fermentation inhibitors during dilute acid hydrolysis of softwood. Enzyme and Microbial Technology, 24(3-4), 151–159.
Palmqvist, E., Grage, H., Meinander, N. Q., & Hahn-Haegerdal, B. (1999). Main and interaction effects of acetic acid, furfural, and p-hydroxybenzoic acid on growth and ethanol productivity of yeasts. Biotechnology and Bioengineering, 63(1), 46–55.
Zaldivar, J., Martinez, A., & Ingram, L. O. (1999). Effect of selected aldehydes on the growth and fermentation of ethanologenic Escherichia coli. Biotechnology and Bioengineering, 65(1), 24–33.
Zeitsch, K. J. (2000). The chemistry and technology of furfural and its many by-products (Vol. 13). Elsevier.
Christian, T. J., Kleiss, B., Yokelson, R. J., Holzinger, R., Crutzen, P. J., Hao, W. M., Saharjo, B. H., & Ward, D. E. (2003). Comprehensive laboratory measurements of biomass-burning emissions: 1. Emissions from Indonesian, African, and other fuels. Journal of Geophysical Research-Atmospheres, 108, D23.
Lemieux, P. M., Lutes, C. C., & Santoianni, D. A. (2004). Emissions of organic air toxics from open burning: a comprehensive review. Progress in Energy and Combustion Science, 30(1), 1–32.
Banerjee, N., Bhatnagar, R., & Viswanathan, L. (1981). Inhibition of glycolysis by furfural in Saccharomyces cerevisiae. European Journal of Applied Microbiology and Biotechnology, 11(4), 226–228.
Modig, T., Liden, G., & Taherzadeh, M. J. (2002). Inhibition effects of furfural on alcohol dehydrogenase, aldehyde dehydrogenase and pyruvate dehydrogenase. The Biochemical Journal, 363(3), 769–776.
Allen, S. A., Clark, W., McCaffery, J. M., Cai, Z., Lanctot, A., Slininger, P. J., Liu, Z. L., & Gorsich, S. W. (2010). Furfural induces reactive oxygen species accumulation and cellular damage in Saccharomyces cerevisiae. Biotechnology for Biofuels, 3(1), 2.
Ask, M., Bettiga, M., Mapelli, V., & Olsson, L. (2013). The influence of HMF and furfural on redox-balance and energy-state of xylose-utilizing Saccharomyces cerevisiae. Biotechnology for Biofuels, 6(1), 22.
Martinez, A., Rodriguez, M. E., Wells, M. L., York, S. W., Preston, J. F., & Ingram, L. O. (2001). Detoxification of dilute acid hydrolysates of lignocellulose with lime. Biotechnology Progress, 17(2), 287–293.
Lee, J. E., Guragain, Y. N., Bastola, K. P., & Vadlani, P. V. (2017). Innovative methods to generate clean sugar stream from biomass feedstocks for efficient fermentation. Bioprocess and Biosystems Engineering, 40(4), 633–641.
Wilson, J. J., Deschatelets, L., & Nishikawa, N. K. (1989). Comparative fermentability of enzymatic and acid hydrolysates of steam-pretreated aspenwood hemicellulose by Pichia stipitis CBS 5776. Applied Microbiology and Biotechnology, 31, 592–596.
López, M. J., Nichols, N. N., Dien, B. S., Moreno, J., & Bothast, R. J. (2004). Isolation of microorganisms for biological detoxification of lignocellulosic hydrolysates. Applied Microbiology and Biotechnology, 64(1), 125–131.
Pienkos, P. T., & Zhang, M. (2009). Role of pretreatment and conditioning processes on toxicity of lignocellulosic biomass hydrolysates. Cellulose., 16(4), 743–762.
Cao, G., Ximenes, E., Nichols, N. N., Zhang, L., & Ladisch, M. (2013). Biological abatement of cellulase inhibitors. Bioresource Technology, 146, 604–610.
Koenig, K., & Andreesen, J. R. (1989). Molybdenum involvement in aerobic degradation of 2-furoic acid by Pseudomonas putida Fu1. Applied and Environmental Microbiology, 55, 1829–1834.
Koopman, F., Wierckx, N., de Winde, J. H., & Ruijssenaars, H. J. (2010). Identification and characterization of the furfural and 5-(hydroxymethyl) furfural degradation pathways of Cupriavidus basilensis HMF14. Proceedings of the National Academy of Sciences of the United States of America, 107(11), 4919–4924.
Lee, S. A., Wrona, L. J., Cahoon, A. B., Crigler, J., Eiteman, M. A., & Altman, E. (2016). Isolation and characterization of bacteria that use furans as the sole carbon source. Applied Biochemistry and Biotechnology, 178(1), 76–90.
Guarnieri, M. T., Franden, M. A., Johnson, C. W., & Beckham, G. T. (2017). Conversion and assimilation of furfural and 5-(hydroxymethyl) furfural by Pseudomonas putida KT2440. Metabolic Engineering Communications, 4, 22–28.
Boopathy, R., Bokang, H., & Daniels, L. (1993). Biotransformation of furfural and 5-hydroxymethyl furfural by enteric bacteria. Journal of Industrial Microbiology, 11(3), 147–150.
Gutiérrez, T., Buszko, M. L., Ingram, L. O., & Preston, J. F. (2002). Reduction of furfural to furfuryl alcohol by ethanologenic strains of bacteria and its effect on ethanol production from xylose. Applied Biochemistry and Biotechnology, 98, 327–340.
Taherzadeh, M. J., Gustafsson, L., Niklasson, C., & Lidén, G. (1999). Conversion of furfural in aerobic and anaerobic batch fermentation of glucose by Saccharomyces cerevisiae. Journal of Bioscience and Bioengineering, 87(2), 169–174.
Tsuge, Y., Hori, Y., Kudou, M., Ishii, J., Hasunuma, T., & Kondo, A. (2014). Detoxification of furfural in Corynebacterium glutamicum under aerobic and anaerobic conditions. Applied Microbiology and Biotechnology, 98(20), 8675–8683.
Gutiérez, T., Ingram, L. O., & Preston, J. F. (2006). Purification and characterization of a furfural reductase (FFR) from Escherichia coli strain LYO1—an enzyme important in the detoxification of furfural during ethanol production. Journal of Biotechnology, 121(2), 154–164.
Liu, Z. L., Moon, J., Andersh, B. J., Slininger, P. J., & Weber, S. (2008). Multiple gene-mediated NAD(P)H-dependent aldehyde reduction is a mechanism of in situ detoxification of furfural and 5-hydroxymethylfurfural by Saccharomyces cerevisiae. Applied Microbiology and Biotechnology, 81(4), 743–753.
Bowman, M. J., Jordan, D. B., Vermillion, K. E., Braker, J. D., Moon, J., & Liu, Z. L. (2010). Stereochemistry of furfural reduction by a Saccharomyces cerevisiae aldehyde reductase that contributes to in situ furfural detoxification. Applied and Environmental Microbiology, 76(15), 4926–4932.
Heux, S., Cachon, R., & Dequin, S. (2006). Cofactor engineering in Saccharomyces cerevisiae: expression of a H2O-forming NADH oxidase and impact on redox metabolism. Metabolic Engineering, 8(4), 303–314.
Heer, D., Heine, D., & Sauer, U. (2009). Resistance of Saccharomyces cerevisiae to high concentrations of furfural is based on NADPH-dependent reduction by at least two oxidoreductases. Applied and Environmental Microbiology, 75(24), 7631–7638.
Trevisan, V. (1889). I generi e le specie delle Batteriacee (pp. 1–35). Zanaboni Gabuzzi.
Casadaban, M. J., & Cohen, S. N. (1980). Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. Journal of Molecular Biology, 138(2), 179–207.
Le Minor, L., & Popoff, M. Y. (1987). Designation of Salmonella enterica sp. nov., nom. rev., as the Type and Only Species of the Genus Salmonella: Request for an Opinion. International Journal of Systematic and Evolutionary Microbiology, 37, 465–468.
Sessitch, A., Coenye, T., Sturz, A. V., Vandamme, P., Barka, E. A., Salles, J. F., Van Elsas, J. D., Faure, D., Reiter, B., Glick, B. R., & Wang-Pruski, G. (2005). Burkholderia phytofirmans sp. nov., a novel plant-associated bacterium with plant-beneficial properties. International Journal of Systematic and Evolutionary Microbiology, 55(3), 1187–1192.
Guyer, M. S., Reed, R. R., Steitz, J. A., & Low, K. B. (1981). Identification of a sex-factor-affinity site in E. coli as γδ, in Cold Spring Harbor symposia on quantitative biology. Cold Spring Harbor Laboratory Press, 45(0), 135–140.
Schroeter, J. (1872). Ueber einige durch Bacterien gebildete Pigmente. Beitr Biol Pflanz, 1, 109–126.
Miller, J. H. (1972). Experiments in molecular genetics. Cold Spring Harbor: Cold Spring Laboratory Press.
Arkin, A. P., Cottingham, R. W., Henry, C. S., Harris, N. L., Stevens, R. L., Maslov, S., Dehal, P., Ware, D., Perez, F., Canon, S., & Sneddon, M. W. (2018). KBase: The United States Department of Energy Systems Biology Knowledgebase. Nature Biotechnology, 36(7), 566–569.
Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., Roop, R. M., & Peterson, K. M. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene., 166(1), 175–176.
Spaink, H. P., Okker, R. J., Wijffelman, C. A., Pees, E., & Lugtenberg, B. J. (1987). Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Molecular Biology, 9(1), 27–39.
Dennis, J. J., & Sokol, P. A. (1995). Electrotransformation of Pseudomonas, in Electroporation protocols for microorganisms (pp. 125–133). Humana Press.
Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. Journal of Molecular Biology, 166(4), 557–580.
Nichols, N. N., & Mertens, J. A. (2008). Identification and transcriptional profiling of Pseudomonas putida genes involved in furoic acid metabolism. FEMS Microbiology Letters, 284(1), 52–57.
Wierckx, N. J., Schuurman, T. D., Kuijper, S. M., & Ruijssenaars, H. J. (2016). Genetically modified cell and process for use of said cell. United States patent 9,309,546.
Werpy, T., Peterson, G., Aden, A., Bozell, J., Holladay, J., White, J., Manheim, A., Elliot, D., Lasure, L., Jones, S., & Gerber, M. (2004). Top value added chemicals from biomass, Volume 1 - Results of screening for potential candidates from sugars and synthesis gas. DOE Scientific and Technical Information. Oak Ridge: US Department of Energy.
Koopman, F., Wierckx, N., de Winde, J. H., & Ruijssenaars, H. J. (2010). Efficient whole-cell biotransformation of 5-(hydroxymethyl) furfural into FDCA, 2, 5-furandicarboxylic acid. Bioresource Technology, 101(16), 6291–6296.
Hossain, G. S., Yuan, H., Li, J., Shin, H. D., Wang, M., Du, G., Chen, J., & Liu, L. (2017). Metabolic engineering of Raoultella ornithinolytica BF60 for production of 2, 5-furandicarboxylic acid from 5-hydroxymethylfurfural. Applied and Environmental Microbiology, 283, e02312–e02316.
Yang, C. F., & Huang, C. R. (2018). Isolation of 5-hydroxymethylfurfural biotransforming bacteria to produce 2, 5-furan dicarboxylic acid in algal acid hydrolysate. Journal of Bioscience and Bioengineering, 125(4), 407–412.
Karich, A., Kleeberg, S. B., Ullrich, R., & Hofrichter, M. (2018). Enzymatic preparation of 2, 5-furandicarboxylic acid (FDCA)—a substitute of terephthalic acid—by the joined action of three fungal enzymes. Microorganisms., 6(1), 5.
Yuan, H., Li, J., Shin, H. D., Du, G., Chen, J., Shi, Z., & Liu, L. (2018). Improved production of 2, 5-furandicarboxylic acid by overexpression of 5-hydroxymethylfurfural oxidase and 5-hydroxymethylfurfural/furfural oxidoreductase in Raoultella ornithinolytica BF60. Bioresource Technology, 247, 1184–1188.
Acknowledgments
We thank Michael E. Kovach for providing the pBBR1MCS-3 vector and Nancy D. Hanson for providing the pMP220 vector.
Funding
This study was funded in part by the Southeastern Sun Grant Center under Prime Award No. DTOS59-07-G-00050.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that there are no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Crigler, J., Eiteman, M.A. & Altman, E. Characterization of the Furfural and 5-Hydroxymethylfurfural (HMF) Metabolic Pathway in the Novel Isolate Pseudomonas putida ALS1267. Appl Biochem Biotechnol 190, 918–930 (2020). https://doi.org/10.1007/s12010-019-03130-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-019-03130-x