Abstract
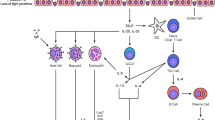
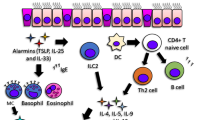
Better management of allergic diseases needs a sharpened understanding of disease heterogeneity and mechanisms in relation to clinically significant outcomes. Phenotypes describing observable clinical and morphologic characteristics and unique responses to treatment have been developed; however, they do not relate to disease mechanisms. Recently, extended heterogeneous and disease-related metabolic, inflammatory, immunological, and remodeling pathways have been described, and reproducible patterns are defined as disease endotypes. An endotype might consist of several intricated mechanisms that cannot be clearly separated into “pure single molecular mechanism” thus being a “complex endotype.” The description of an endotype may rely on biomarkers, which can be the signature of a complex underlying pathway or a key molecule associated with or directly playing a role in a particular disease endotype. The Th2 type inflammation can be defined as a complex endotype in asthma and linked to mechanisms of disease development and response to treatment and to disease outcomes such as exacerbations and remodeling. The type 2 complex endotype in allergies and asthma includes innate lymphoid cells, T helper 2 cells, tissue eosinophilia, and IgE production. Currently, emerging endotype-driven strategies in asthma, particularly the development of biologicals that target a single molecular pathway, are being focused for solving individualized clinical problems on disease outcomes. Progress is also being made for endotyping rhinitis, chronic rhinosinusitis, and atopic dermatitis.


Similar content being viewed by others
Abbreviations
- ADMA:
-
Asymmetric dimethylarginine
- CLC:
-
Charcot-Leyden crystal protein
- CPA3:
-
Carboxypeptidase A3
- DNA-SE1L3:
-
Deoxyribonuclease I-like 3
- DC:
-
Dendritic cells
- Eos:
-
Eosinophils
- FeNO:
-
Fractional exhaled NO
- ICS:
-
Inhaled corticosteroids
- ILC:
-
Innate lymphoid cells
- l-Arg:
-
l-Arginine
- NKT cells:
-
Natural killer T cells
- PG:
-
Prostaglandin
- SARP:
-
Severe Asthma Respiratory Program
- Th:
-
T helper cell
- TSLP:
-
Thymic stromal lymphopoietin
- VOCs:
-
Volatile organic compounds
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Akdis CA. Therapies for allergic inflammation: refining strategies to induce tolerance. Nat Med. 2012;18:736–49.
Akdis M, Akdis C. Immune tolerance. In: Bochner BS, Adkinson Jr NF, Burks W, Busse WW, Holgate Jr ST, Lemanski RF, O’Hehir RE, editors. Middleton’s allergy. 8th ed. Cambridge: Elsevier; 2013.
Akdis M et al. Mechanisms of allergen-specific immunotherapy: multiple suppressor factors at work in immune tolerance to allergens. J Allergy Clin Immunol. 2014;133:621–31.
Pillai RA, Calhoun WJ. Introduction to asthma and phenotyping. Adv Exp Med Biol. 2014;795:5–15.
Lotvall J et al. Asthma endotypes: a new approach to classification of disease entities within the asthma syndrome. J Allergy Clin Immunol. 2011;127:355–60. The PRACTALL consensus was the first to propose the criteria to validate an asthma endotype.
Agache IO. From phenotypes to endotypes to asthma treatment. Curr Opin Allergy Clin Immunol. 2013;13:249–56. The review critically revises new approaches to classify asthma and the emerging endotype-driven strategies.
Agache I et al. Untangling asthma phenotypes and endotypes. Allergy. 2012;67:835–46. Comprehensive description of asthma phenotypes and corresponding endotypes grading the level of evidence for corresponding biomarkers.
Agache IO. Endotype driven treatment of asthma. Curr Treat Options Allergy. 2014;1:198–212.
Scanlon ST et al. Type 2 innate lymphoid cells: new players in asthma and allergy. Curr Opin Immunol. 2012;24:707–12.
Holgate ST. Innate and adaptive immune responses in asthma. Nat Med. 2012;18:673–83.
DeKruyff RH et al. Innate immunity in the lung regulates the development of asthma. Immunol Rev. 2014;260:235–48.
Akbari O et al. CD4+ invariant T-cell-receptor+ natural killer T cells in bronchial asthma. N Engl J Med. 2006;354:1117–29.
Gauvreau GM et al. Effects of an anti-TSLP antibody on allergen-induced asthmatic responses. N Engl J Med. 2014;370:2102–10.
Dua B et al. Myeloid dendritic cells type 2 in allergic asthma. Allergy. 2013;68:1322–6.
Melum GR, et al. A thymic stromal lymphopoietin-responsive dendritic cell subset mediates allergic responses in the upper airway mucosa. J Allergy Clin Immunol. 2014;134(3):613–21
Jacobsen EA, et al. Re-defining the unique roles for eosinophils in allergic respiratory inflammation. Clin Exp Allergy. 2014;44(9):1119–36
Kim K et al. Comparative analysis of human epidermal and peripheral blood gammadelta T cell cytokine profiles. Ann Dermatol. 2014;26:308–13.
Kabata H et al. Thymic stromal lymphopoietin induces corticosteroid resistance in natural helper cells during airway inflammation. Nat Commun. 2013;4:2675.
Kim HY et al. Innate lymphoid cells responding to IL-33 mediate airway hyperreactivity independently of adaptive immunity. J Allergy Clin Immunol. 2012;129:216–27. e1-6.
Kondo Y et al. Administration of IL-33 induces airway hyperresponsiveness and goblet cell hyperplasia in the lungs in the absence of adaptive immune system. Int Immunol. 2008;20:791–800.
Gold MJ et al. Group 2 innate lymphoid cells facilitate sensitization to local, but not systemic, TH2-inducing allergen exposures. J Allergy Clin Immunol. 2014;133:1142–8.
Halim TY et al. Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation. Immunity. 2014;40:425–35.
Mjosberg JM et al. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat Immunol. 2011;12:1055–62. This paper describe human type 2 innate lymphoid cells for the first time.
Kim BS et al. TSLP elicits IL-33-independent innate lymphoid cell responses to promote skin inflammation. Sci Transl Med. 2013;5:170ra16.
Teunissen MB, et al. Composition of innate lymphoid cell subsets in the human skin: enrichment of NCR ILC3 in lesional skin and blood of psoriasis patients. J Invest Dermatol. 2014;134(9):2351–60
Allakhverdi Z et al. CD34+ hemopoietic progenitor cells are potent effectors of allergic inflammation. J Allergy Clin Immunol. 2009;123:472–8.
Shikotra A et al. Increased expression of immunoreactive thymic stromal lymphopoietin in patients with severe asthma. J Allergy Clin Immunol. 2012;129:104–11. e1-9.
Shaw JL et al. IL-33-responsive innate lymphoid cells are an important source of IL-13 in chronic rhinosinusitis with nasal polyps. Am J Respir Crit Care Med. 2013;188:432–9.
Nagarkar DR et al. Thymic stromal lymphopoietin activity is increased in nasal polyps of patients with chronic rhinosinusitis. J Allergy Clin Immunol. 2013;132:593–600. e12.
Kamekura R et al. The role of IL-33 and its receptor ST2 in human nasal epithelium with allergic rhinitis. Clin Exp Allergy. 2012;42:218–28.
Gervais FG et al. Selective modulation of chemokinesis, degranulation, and apoptosis in eosinophils through the PGD2 receptors CRTH2 and DP. J Allergy Clin Immunol. 2001;108:982–8.
Pettipher R et al. Heightened response of eosinophilic asthmatic patients to the CRTH2 antagonist OC000459. Allergy. 2014;69:1223–32.
Hirai H et al. Prostaglandin D2 selectively induces chemotaxis in T helper type 2 cells, eosinophils, and basophils via seven-transmembrane receptor CRTH2. J Exp Med. 2001;193:255–61.
Xue L et al. Prostaglandin D2 activates group 2 innate lymphoid cells through chemoattractant receptor-homologous molecule expressed on TH2 cells. J Allergy Clin Immunol. 2014;133:1184–94.
Doherty TA et al. Allergen challenge in allergic rhinitis rapidly induces increased peripheral blood type 2 innate lymphoid cells that express CD84. J Allergy Clin Immunol. 2014;133:1203–5.
Dougherty RH et al. Accumulation of intraepithelial mast cells with a unique protease phenotype in T(H)2-high asthma. J Allergy Clin Immunol. 2010;125:1046–53. e8.
Oh CK et al. Biology of the interleukin-9 pathway and its therapeutic potential for the treatment of asthma. Inflamm Allergy Drug Targets. 2011;10:180–6.
Fajt ML et al. Prostaglandin D(2) pathway upregulation: relation to asthma severity, control, and TH2 inflammation. J Allergy Clin Immunol. 2013;131:1504–12.
Barnes N et al. A randomized, double-blind, placebo-controlled study of the CRTH2 antagonist OC000459 in moderate persistent asthma. Clin Exp Allergy. 2012;42:38–48. Prood-of-concept study testing the safety and efficacy of targeted PGD2 intervention in Th2 high asthma.
Steinke JW et al. Prominent role of IFN-gamma in patients with aspirin-exacerbated respiratory disease. J Allergy Clin Immunol. 2013;132:856–65. e1-3.
Rebane A et al. Mechanisms of IFN-gamma-induced apoptosis of human skin keratinocytes in patients with atopic dermatitis. J Allergy Clin Immunol. 2012;129:1297–306.
Wenzel SE et al. Asthmatic granulomatosis: a novel disease with asthmatic and granulomatous features. Am J Respir Crit Care Med. 2012;186:501–7.
Linden A, et al. Interleukin-17 cytokine signalling in patients with asthma. Eur Respir J. 2014;44(5):1319–31
Agache I et al. Increased serum IL-17 is an independent risk factor for severe asthma. Respir Med. 2010;104:1131–7.
Szczeklik A et al. The broken balance in aspirin hypersensitivity. Eur J Pharmacol. 2006;533:145–55.
Desai D et al. Elevated sputum interleukin-5 and submucosal eosinophilia in obese individuals with severe asthma. Am J Respir Crit Care Med. 2013;188:657–63.
Holguin F et al. An association between l-arginine/asymmetric dimethyl arginine balance, obesity, and the age of asthma onset phenotype. Am J Respir Crit Care Med. 2013;187:153–9. The study advances a new hypothesis of metabolic disturbed pathway in obese asthma.
Conus S et al. Leptin is an eosinophil survival factor. J Allergy Clin Immunol. 2005;116:1228–34.
Wong CK et al. Leptin-mediated cytokine release and migration of eosinophils: implications for immunopathophysiology of allergic inflammation. Eur J Immunol. 2007;37:2337–48.
Han NR, et al. TSLP induces mast cell development and aggravates allergic reactions through the activation of MDM2 and STAT6. J Invest Dermatol. 2014;134(10):2521–30.
Soyka MB et al. Defective epithelial barrier in chronic rhinosinusitis: the regulation of tight junctions by IFN-gamma and IL-4. J Allergy Clin Immunol. 2012;130:1087–96. e10.
Woodruff PG et al. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med. 2009;180:388–95. First in vivo study validating the concept of Th2 high and low asthma in relation to response to treatment and to lung remodelling.
Green RH, Brightling CE, McKenna S, et al. Asthma exacerbations and sputum eosinophil counts: a randomised controlled trial. Lancet 2002;360:1715–21.
Pijnenburg MW, De Jongste JC. Exhaled nitric oxide in childhood asthma: a review. Clin Exp Allergy. 2008;38:246–59.
Petsky HL, Cates CJ, Li A, et al. Tailored interventions based on exhaled nitric oxide versus clinical symptoms for asthma in children and adults. Cochrane Database Syst Rev. 2009;4, CD006340.
Kanemitsu Y, Matsumoto H, Mishima M, et al. Factors contributing to an accelerated decline in pulmonary function in asthma. Allergol Int. 2014;63:181–8.
Tran TN, Khatry DB, Ke X, et al. High blood eosinophil count is associated with more frequent asthma attacks in asthma patients. Ann Allergy Asthma Immunol. 2014;113(1):19–24.
ten Brinke A, Zwinderman AH, Sterk PJ et al. Factors associated with persistent airflow limitation in severe asthma. Am J Respir Crit Care Med. 2001;164:744–8.
Peters MC, Mekonnen ZK, Yuan S, et al. Measures of gene expression in sputum cells can identify TH2-high and TH2-low subtypes of asthma. J Allergy Clin Immunol. 2014;133:388–94.
Baines KJ, Simpson JL, Wood LG, et al. Sputum gene expression signature of 6 biomarkers discriminates asthma inflammatory phenotypes. J Allergy Clin Immunol 2014;133:997–1007.
Poole A, Urbanek C, Eng C, et al. Dissecting childhood asthma with nasal transcriptomics distinguishes subphenotypes of disease. J Allergy Clin Immunol. 2014;133(3):670–8
Little FF, Delgado DM, Wexler PJ, et al. Salivary inflammatory mediator profiling and correlation to clinical disease markers in asthma. PLoS One. 2014;9(1):e84449.
Hanania NA, Wenzel S, Rosén K, et al. Exploring the effects of omalizumab in allergic asthma: an analysis of biomarkers in the EXTRA study. Am J Respir Crit Care Med. 2013;187:804–11. First study to validate a composite biomarker measure for effectiveness of omalizumab in severe allergic asthma.
Ortega H, Li H, Suruki R, et al. Cluster analysis and characterization of response to mepolizumab: a step closer to personalized medicine for patients with severe asthma. Ann Am Thorac Soc. 2014 Jul 1. [Epub ahead of print].
Agache I, Ciobanu C. Persistent high FeNO phenotype in asthma. J Allergy Clin Immunol. 2011;127:AB4.
Agache I, Ciobanu C. Predictive value of lung function trend and FeNO for difficult asthma in children. J Investig Allergol Clin Immunol 2012;22:419–26.
Stern G, de Jongste J, van der Valk R, et al. Fluctuation phenotyping based on daily fraction of exhaled nitric oxide values in asthmatic children. J Allergy Clin Immunol 2011;128:293–300.
Newby C, Agbetile J, Hargadon B, Monteiro W, Green R, Pavord I et al. Lung function decline and variable airway inflammatory pattern: longitudinal analysis of severe asthma. J Allergy Clin Immunol. 2014;134(2):287–94.
van der Schee MP, Palmay R, Cowan JO, et al. Predicting steroid responsiveness in patients with asthma using exhaled breath profiling. Clin Exp Allergy. 2013;43:1217–25.
Malinovschi A et al. Exhaled nitric oxide levels and blood eosinophil counts independently associate with wheeze and asthma events in National Health and Nutrition Examination Survey subjects. J Allergy Clin Immunol. 2013;132:821–7. e1-5. The study shows that blood eosinophils and FeNO are independent predictors for asthma events and response to treatment supporting the 2 main sub-endotypes of Th2 inflammation.
Pavord ID et al. Mepolizumab for severe eosinophilic asthma (DREAM): a multicentre, double-blind, placebo-controlled trial. Lancet. 2012;380:651–9. DREAM is the first large randomized study validating the endotype-driven approach in severe asthma as a method to increase response to targeted treatment (anti IL-5). Patients were selected based on a composite measure of eosinophilic inflammation.
Corren J et al. Lebrikizumab treatment in adults with asthma. N Engl J Med. 2011;365:1088–98. MILLY is one of the first targeted treatments in asthma highlighting the value of a biomarker used as a pathway specific diagnostic test.
Jia G et al. Periostin is a systemic biomarker of eosinophilic airway inflammation in asthmatic patients. J Allergy Clin Immunol. 2012;130:647–54. e10. The BOBCAT data prove the value of serum periostin as a reliable and noninvasive biomarker of eosinophilic inflammation in asthma.
Van Zele T et al. Differences in initial immunoprofiles between recurrent and nonrecurrent chronic rhinosinusitis with nasal polyps. Am J Rhinol Allergy. 2014;28:192–8.
Akdis CA et al. Endotypes and phenotypes of chronic rhinosinusitis: a PRACTALL document of the European Academy of Allergy and Clinical Immunology and the American Academy of Allergy, Asthma & Immunology. J Allergy Clin Immunol. 2013;131:1479–90. The PRACTALL consensus evaluates the relation between the CRS endotypes and disease severity and response to treatment.
Mu Z, et al. Molecular biology of atopic dermatitis. Clin Rev Allergy Immunol. 2014.
Wollenberg A et al. Immunological and molecular targets of atopic dermatitis treatment. Br J Dermatol. 2014;170 Suppl 1:7–11.
Compliance with Ethics Guidelines
Conflict of Interest
Ioana Agache, Kazunari Sugita, Hideaki Morita, Mübeccel Akdis, and Cezmi A. Akdis declare that they have no conflicts of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Topical Collection on Asthma
Rights and permissions
About this article
Cite this article
Agache, I., Sugita, K., Morita, H. et al. The Complex Type 2 Endotype in Allergy and Asthma: From Laboratory to Bedside. Curr Allergy Asthma Rep 15, 29 (2015). https://doi.org/10.1007/s11882-015-0529-x
Published:
DOI: https://doi.org/10.1007/s11882-015-0529-x