Abstract
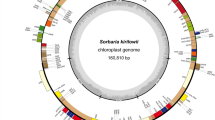
Retrophyllum piresii (Podocarpaceae) is an endemic conifer species from the Brazilian Amazonian Region, and very few data related to ecological and genetic characteristics of this species are available. Plastome sequencing is an efficient tool to understand enigmatic and basal phylogenetic relationships at different taxonomic levels, as well as to probe the structural and functional evolution of plants. Usually, the plastome of photosynthetic land plants is quadripartite, with two copies of the inverted repeats (IRs) separating the small and large single-copy regions. However, in gymnosperms, IR can vary from large in size to completely absent, being constituted principally by transfer RNA (tRNA) genes, or a part of sequence of other genes. Here, we sequenced and characterized the complete plastome of R. piresii. This plastome was determined to be 133,291 bp (~480-fold coverage), presenting a total of 120 identified genes, of which 118 were single copy and two genes, trnN-GUU and trnD-GUC, were found to be duplicated and occurring as inverted and directed repeat (DR) sequences, respectively. These repeated regions presented recombinationally active sites, resulting in an IR-mediated inversion and a DR-mediated deletion. However, the isoform resulted from DR-mediated deletion may result in unviable plastome, with deletion of photosynthetic and expression machinery-related genes.



Similar content being viewed by others
References
Ahmed I, Matthews PJ, Biggs PJ, Naeem M, McLenachan PA, Lockhart PJ (2013) Identification of chloroplast genome loci suitable for high-resolution phylogeographic studies of Colocasia esculenta (L.) Schott (Araceae) and closely related taxa. Mol Ecol Resour 13:929–937
Alkatib S, Scharff LB, Rogalski M, Fleischmann TT, Matthes A et al (2012) The contributions of wobbling and superwobbling to the reading of the genetic code. PLoS Genet 8(11), e1003076
Baskauf CJ, Jinks NC, Mandel JR, McCauley DE (2014) Population genetics of Braun’s Rockcress (Boechera perstellata, Brassicaceae), an endangered plant with a disjunct distribution. J Hered 105:265–275
Besnard G, Hernández P, Khadari B, Dorado G, Savolainen V (2011) Genomic profiling of plastid DNA variation in the Mediterranean olive tree. BMC Plant Biol 11:80
Biffin E, Conran J, Lowe A (2011) Podocarp evolution: a molecular phylogenetic perspective. In: Turner BL, Cernusak LA (eds) Ecology of the Podocarpaceae in tropical forests. Smithsonian Institution Scholarly Press, Washington, pp 1–20
Cronn R, Liston A, Parks M, Gernandt DS, Shen R et al (2008) Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Res 36, e122
Day A, Kode V, Madesis P, Iamtham S (2005) Simple and efficient removal of marker genes from plastids by homologous recombination. Methods Mol Biol 286:255–270
Delplancke M, Alvarez N, Espíndola A, Joly H, Benoit L, Brouck E, Arrigo N (2012) Gene flow among wild and domesticated almond species: insights from chloroplast and nuclear markers. Evol Appl 5:317–329
Dexter KG, Terborgh JW, Cunningham CW (2012) Historical effects on beta diversity and community assembly in Amazonian trees. Proc Natl Acad Sci U S A 109:7787–7792
Do HDK, Kim JS, Kim J (2014) A trnI_CAU triplication event in the complete chloroplast genome of Paris verticillata M.Bieb. (Melanthiaceae, Liliales). Genome Biol Evol 6:1699–1706
Farjon A (1998) World checklist and bibliography of conifers. The Royal Botanical Gardens, Kew
Fleischmann TT, Scharff LB, Alkatib S, Hasdorf S, Schottler MA et al (2011) Nonessential plastid-encoded ribosomal proteins in tobacco: a developmental role for plastid translation and implications for reductive genome evolution. Plant Cell 23:3137–3155
Gray BN, Ahner BA, Hanson MR (2009) Extensive homologous recombination between introduced and native regulatory plastid DNA elements in transplastomic plants. Transgenic Res 18:559–572
Guo X, Castillo-Ramírez S, González V, Bustos P, Fernández-Vázquez JL, Santamaría RI, Arellano J, Cevallos MA, Dávila G (2007) Rapid evolutionary change of common bean (Phaseolus vulgaris L) plastome, and the genomic diversification of legume chloroplasts. BMC Genomics 8:228
Guo W, Grewe F, Cobo-Clark A, Fan W, Duan Z, Adams RP, Schwarzbach AE, Mower JP (2014) Predominant and substoichiometric isomers of the plastid genome coexist within Juniperus plants and have shifted multiple times during cupressophyte evolution. Genome Biol Evol 6:580–590
Gurdon C, Maliga P (2014) Two distinct plastid genome configurations and unprecedented intraspecies length variation in the accD coding region in Medicago truncatula. DNA Res 21:417–427
Hansen DR, Dastidar SG, Cai Z, Penaflor C, Kuehl JV, Boore JL, Jansen RK (2007) Phylogenetic and evolutionary implications of complete chloroplast genome sequences of four early-diverging angiosperms: Buxus (Buxaceae), Chloranthus (Chloranthaceae), Dioscorea (Dioscoreaceae), and Illicium (Schisandraceae). Mol Phylogenet Evol 45:547–563
Hirao T, Watanabe A, Kurita M, Kondo T, Takata K (2008) Complete nucleotide sequence of the Cryptomeria japonica D. Don. chloroplast genome and comparative chloroplast genomics: diversified genomic structure of coniferous species. BMC Plant Biol 8:70
Iamtham S, Day A (2000) Removal of antibiotic resistance genes from transgenic tobacco plastids. Nat Biotechnol 18:1172–1176
Jansen RK, Ruhlman TA (2012) Plastid genomes of seed plants. In: Bock R, Knoop V (eds) Genomics of chloroplasts and mitochondria. Springer, Netherlands, pp 103–126
Jansen RK, Cai Z, Raubeson LA, Daniell H, dePamphilis CW et al (2007) Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci U S A 104:19369–19374
Kato S, Iwata H, Tsumura Y, Mukai Y (2011) Genetic structure of island populations of Prunus lannesiana var. speciosa revealed by chloroplast DNA, AFLP and nuclear SSR loci analyses. J Plant Res 124:11–23
Kato S, Imai A, Rie N, Mukai Y (2013) Population genetic structure in a threatened tree, Pyrus calleryana var. dimorphophylla revealed by chloroplast DNA and nuclear SSR locus polymorphisms. Conserv Genet 14:983–996
Knox EB (2014) The dynamic history of plastid genomes in the Campanulaceae sensu lato is unique among angiosperms. Proc Natl Acad Sci U S A 111:11097–11102
Kode V, Mudd EA, Iamtham S, Day A (2006) Isolation of precise plastid deletion mutants by homology-based excision: a resource for site-directed mutagenesis, multi-gene changes and high-throughput plastid transformation. Plant J 46:901–909
Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R (2001) REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res 29:4633–4642
Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL (2004) Versatile and open software for comparing large genomes. Genome Biol 5:R12
Lin C, Huang J, Wu C, Hsu C, Chaw S (2010) Comparative chloroplast genomics reveals the evolution of Pinaceae genera and subfamilies. Genome Biol Evol 2:504–517
Lin CP, Wu C, Huang Y, Chaw S (2012) The complete chloroplast genome of Ginkgo biloba reveals the mechanism of inverted repeat contraction. Genome Biol Evol 4:374–381
Lohse M, Drechsel O, Kahlau S, Bock R (2013) OrganellarGenomeDRAW: a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucl Acids Res. doi:10.1093/nar/gkt289
López P, Tremetsberger K, Kohl G, Stuessy T (2012) Progenitor-derivative speciation in Pozoa (Apiaceae, Azorelloideae) of the southern Andes. Ann Bot 109:351–363
Moore MJ, Bell CD, Soltis PS, Soltis DE (2007) Using plastid genomic-scale data to resolve enigmatic relationships among basal angiosperms. Proc Natl Acad Sci U S A 104:19363–19368
Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE (2010) Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc Natl Acad Sci U S A 107:4623–4628
Murray BG (2013) Karyotype variation and evolution in gymnosperms. In: Leitch IJ, Greilhuber J, Dolezel J, Wendel JF (eds) Plant genome diversity 2: physical structure, behaviour and evolution of plant genomes. Springer, Vienna, p 231–243
Palmer JD (1983) Chloroplast DNA exists in two orientations. Nature 301:92–93
Parks M, Cronn R, Liston A (2009) Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol 7:84
Powell W, Morgante M, Andre C, McNicol JW, Machray GC, Doyle JJ, Tingey SV, Rafalski JA (1995) Hypervariable microsatellites provide a general source of polymorphic DNA markers for the chloroplast genome. Curr Biol 5:1023–1029
Provan J, Powell W, Hollingsworth PM (2001) Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trends Ecol Evol 16:142–147
Rogalski M, Ruf S, Bock R (2006) Tobacco plastid ribosomal protein S18 is essential for cell survival. Nucleic Acids Res 34:4537–4545
Rogalski M, Schottler MA, Thiele W, Schulze WX, Bock R (2008a) Rpl33, a nonessential plastid-encoded ribosomal protein in tobacco, is required under cold stress conditions. The Plant Cell 20:2221–2237
Rogalski M, Karcher D, Bock R (2008b) Superwobbling facilitates translation with reduced tRNA sets. Nat Struct Mol Biol 15:192–198
Rogalski M, Vieira LN, Fraga HPF, Guerra MP (2015) Plastid genomics in horticultural species: importance and applications for plant population genetics, evolution, and biotechnology. Front Plant Sci 6:586
Roullier C, Rossel G, Tay D, McKey D, Lebot V (2011) Combining chloroplast and nuclear microsatellites to investigate origin and dispersal of New World sweet potato landraces. Mol Ecol 20:3963–3977
Roullier C, Duputié A, Benoit L, Fernández-Bringas VM et al (2013) Correction: disentangling the origins of cultivated sweet potato (Ipomoea batatas (L.) Lam). PLoS One 8(10):10.1371
Ruhsam M, Rai HS, Mathews S, Ross TG, Graham SW et al (2015) Does complete plastid genome sequencing improve species discrimination and phylogenetic resolution in Araucaria? Mol Ecol Resour 15:1067–1078
Schattner P, Brooks AN, Lowe TM (2005) The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33:W686–W689
Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N et al (1986) The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J 5:2043–2049
Stein DB, Palmer JD, Thompson WF (1986) Structural evolution and flip-flop recombination of chloroplast DNA in the fern genus Osmunda. Curr Genet 10:835–841
Tangphatsornruang S, Sangsrakru D, Chanprasert J, Uthaipaisanwong P, Yoocha T et al (2009) The chloroplast genome sequence of mungbean (Vigna radiata) determined by high-throughput pyrosequencing: structural organization and phylogenetic relationships. DNA Res 17:11–22
Tomar RSS, Deshmukh RK, Naik BK, Tomar SMS, Vinod (2014) Development of chloroplast-specific microsatellite markers for molecular characterization of alloplasmic lines and phylogenetic analysis in wheat. Plant Breed 133:12–18
Tsudzuki J, Nakashima K, Tsudzuki T, Hiratsuka J, Shibata M et al (1992) Chloroplast DNA of black pine retains a residual inverted repeat lacking rRNA genes: nucleotide sequences of trnQ, trnK, psbA, trnI and trnH and the absence of rps16. Mol Gen Genet 232:206–214
Vendramin GG, Anzidei M, Madaghiele A, Bucci G (1998) Distribution of genetic diversity in Pinus pinaster Ait. as revealed by chloroplast microsatellites. Theor Appl Genet 97:456–463
Vieira LN, Faoro H, Rogalski M, Fraga HPF, Cardoso RLA (2014a) The complete chloroplast genome sequence of Podocarpus lambertii: genome structure, evolutionary aspects, gene content and SSR detection. PLoS One 9(3), e90618
Vieira LN, Faoro H, Fraga HPF, Rogalski M, de Souza EM et al (2014b) An improved protocol for intact chloroplasts and cpDNA isolation in conifers. PLoS One 9(1), e84792
Wakasugi T, Tsudzuki J, Ito S, Nakashima K, Tsudzuki T et al (1994) Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. Proc Natl Acad Sci U S A 91:9794–9798
Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D (2011) The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol 76:273–297
Wu CS, Chaw SM (2014) Highly rearranged and size-variable chloroplast genomes in conifers II clade (cupressophytes): evolution towards shorter intergenic spacers. Plant Biotechnol J doi. doi:10.1111/pbi.12141
Wu CS, Wang YN, Liu SM, Chaw SM (2007) Chloroplast genome (cpDNA) of Cycas taitungensis and 56 cp protein-coding genes of Gnetum parvifolium: insights into cpDNA evolution and phylogeny of extant seed plants. Mol Biol Evol 24:1366–1379
Wu CS, Lai YT, Lin CP, Wang YN, Chaw SM (2009) Evolution of reduced and compact chloroplast genomes (cpDNAs) in gnetophytes: selection towards a lower cost strategy. Mol Phylogent Evol 52:115–124
Wu CS, Wang YN, Hsu CY, Lin CP, Chaw SM (2011) Loss of different inverted repeat copies from the chloroplast genomes of Pinaceae and cupressophytes and influence of heterotachy on the evaluation of gymnosperm phylogeny. Genome Biol Evol 3:1284–1295
Wyman SK, Jansen RK, Boore JL (2004) Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3252–3255
Yi X, Gao L, Wang B, Su Y, Wang T (2013) The complete chloroplast genome sequence of Cephalotaxus oliveri (Cephalotaxaceae): evolutionary comparison of cephalotaxus chloroplast DNAs and insights into the loss of inverted repeat copies in gymnosperms. Genome Biol Evol 5:688–698
Zhang Y, Ma J, Yang B, Li R, Zhu W, Sun L, Tian J, Zhang L (2014) The complete chloroplast genome sequence of Taxus chinensis var. mairei (Taxaceae): loss of an inverted repeat region and comparative analysis with related species. Gene 540:201–209
Acknowledgments
This work was supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), and Fundação de Amparo à Pesquisa e Inovação do Estado de Santa Catarina (FAPESC 14848/2011-2, 3770/2012, and 2780/2012-4). The authors thank Prof. Dr. Willian Antônio Rodrigues for the information related to the species, Angelo Schuabb Heringer and Ramon Felipe Scherer for collecting the plant material, and Museu Paraense Emílio Goeldi for kindly providing the plant material.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Y. Tsumura
Leila do Nascimento Vieira and Marcelo Rogalski contributed equally to this work.
Rights and permissions
About this article
Cite this article
do Nascimento Vieira, L., Rogalski, M., Faoro, H. et al. The plastome sequence of the endemic Amazonian conifer, Retrophyllum piresii (Silba) C.N.Page, reveals different recombination events and plastome isoforms. Tree Genetics & Genomes 12, 10 (2016). https://doi.org/10.1007/s11295-016-0968-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-016-0968-0