Abstract
Even though complexity is a concept that is ubiquitously used by biologists and philosophers of biology, it is rarely made precise. I argue that a clarification of the concept is neither trivial nor unachievable, and I propose a unifying framework based on the technical notion of “effective complexity” that allows me to do justice to conflicting intuitions about biological complexity, while taking into account several distinctions in the usage of the concept that are often overlooked. In particular, I propose a distinction between two kinds of complexity, “mechanical” and “emergent”, which can be understood as different ways of relating the effective complexity of mechanisms and of behaviors in biological explanations. I illustrate the adequacy of this framework by discussing different attempts to understand intracellular organization in terms of pathways and networks. My framework provides a different way of thinking about recent philosophical debates, for example, on the difference between mechanistic and topological explanations and about the concept of emergence. Moreover, it can contribute to a proper assessment of metascientific arguments that invoke biological complexity.
Similar content being viewed by others
1 Introduction
Everybody agrees that living systems are complex. An altogether different question, however, is whether everybody agrees about what they actually mean by “complex”. This question is not easy to answer because when using the term, biologists mostly do not engage in much further reflection on its meaning. One exception is the debate around the controversial question of whether evolution shows a trend of increasing complexity (e.g. Bonner 1988; Smith and Szathmary 1995; Gould 1996). In order to understand whether complexity changes across episodes of evolutionary change, evolutionary biologists have tried to come up with quantitative measures for the complexity of an organism, or of some of its structural features (e.g. McShea 2005). Yet, even in this context, as one of the involved scientists soberly remarks, “[m]ost agree ...that nobody knows precisely what is meant by the word ‘complexity’ when referring to a biological organism” (Adami 2002, p. 1085).
Complexity is not untypical in this regard, of course. Biologists are mainly concerned with using concepts and not with clarifying or defining them. So if we want to get a clearer picture, we should perhaps consult the work of philosophers, one of whose core businesses is, after all, the analysis of concepts. It turns out, however, that here we are faced with a similar problem. Just like biologists themselves, philosophers of biology frequently talk about complexity, but they typically do not make explicit, or are at least not very precise about, what they mean by it. One reason could be that complexity is simply a rather trivial concept and clarification not worth any serious effort. Alternatively, further clarification might be impossible because the concept itself is too complex.
In this paper, I argue that further clarification of the notion of biological complexity is an important, non-trivial, and achievable task. I propose a unifying framework based on Murray Gell-Mann’s notion of “effective complexity” that allows me to make sense of different and partially conflicting intuitions about biological complexity and to take into account some distinctions in the usage of the concept that are often overlooked in the philosophical literature. In particular, I argue that in the context of biology “complexity” is both used with reference to the behavior of a system and with reference to the underlying mechanism. Different perspectives on complexity can be understood as different ways of relating these two uses of the term. My illustrations are mainly taken from the fields of molecular and cell biology because they provide particularly instructive examples, but I think that the basic concepts can be applied to biology more generally.
The paper is structured as follows. In Sect. 2, I motivate the claim that the concept of biological complexity needs further clarification. Section 3 discusses a number of distinctions that a clarified account of complexity must take into consideration. Section 4 outlines my unified perspective on biological complexity, while Sect. 5 shows the adequacy of this perspective by applying it to concrete examples of research in molecular and cell biology. I conclude with some remarks on the relevance of my framework for recent debates in philosophy of biology.
2 Ambiguity and vagueness around biological complexity
In two seminal works on biological complexity, Bechtel and Richardson’s Discovering Complexity and Sandra Mitchell’s Biological Complexity and Integrative Pluralism, one finds surprisingly little explicit discussion of the meaning of “complexity”. Bechtel and Richardson, for example, write that
[m]any machines are simple, consisting of only a handful of parts that interact minimally or in a linear way. In these machines we can trace and describe the events occurring straightforwardly, relating first what is done by one component, then how this affects the next. Such machines induce little cognitive strain. Some machines, however, are much more complex: one component may affect and be affected by several others, with a cascading effect; or there may be significant feedback from “later” to “earlier” stages. In the latter case, what is functionally dependent becomes unclear. Interaction among components becomes critical. Mechanisms of this latter kind are complex systems. In the extreme they are integrated systems. In such cases, attempting to understand the operation of the entire machine by following the activities in each component in a brute force manner is liable to be futile. (Bechtel and Richardson 2010, p. 18)
And the closest Mitchell comes to something like a general definition is in the following passage:
Minimally, complex systems can be distinguished from simple objects by having multiple parts that stand in nonsimple relations. That is, there is structure or order in the way in which the whole is composed of the parts. (Mitchell 2003, p. 5)
While highly suggestive, these characterizations raise more questions than answers. What does it mean, for example, that interactions in a system are critical? What is a nonsimple relation? When do we say that there is structure or order in a system? Thus, they are hardly sufficient guides when it comes to determining whether, or in what way a particular system is complex, or when we want to compare the complexity of different systems.
Is this a problem? Perhaps not. Perhaps the concept of biological complexity is intuitively clear enough, or even trivial, and any attempt at further clarification would amount to the superfluous exercise of spelling out what everybody already knows. Conversely, one may think that attempts of further clarifying complexity are futile because the notion itself is too complex. Perhaps it simply does not allow for one unifying definition but requires many different accounts depending on the context. A third possibility is that nothing much hinges on the exact meaning of the term. Biologists and philosophers may often use it, but it may in most contexts not play an important theoretical role. Perhaps complexity, just like the concept of life itself, refers to a feature that is so fundamental in biology that it rarely comes to the fore?
The first option, according to which an intuitive notion is clear enough, does not seem plausible. In particular, it would imply that Bechtel and Richardson and Mitchell allude to the same “simple” idea of complexity. But when considering the quotes above, one can already see that potentially diverging intuitions are involved. While Bechtel and Richardson emphasize the fact that complex systems resist simple description, Mitchell takes order to be a hallmark of complexity, and thus a feature that prima facie makes our attempts at describing and understanding a system easier. A clarified account of complexity would minimally need to spell out how one should balance the idea of complexity as related to structure or order against the idea of complexity as “complicatedness”.
The second option, according to which a unified idea of complexity is not available because the concept itself is too heterogeneous, seems to be what Sandra Mitchell has in mind:
The multiplicity of definitions of “complexity” reflects not confusion on the part of scientists but the actual variety of ways that systems are complex. (Mitchell 2003, p. 4)
Mitchell is specifically referring to the observation that while many different measures of complexity have been proposed for various purposes, no single measure seems to work in every context (Lloyd and Pagels 1988). A similar view on the “complexity of complexity” has been articulated by Rescher (1998). In response to this observation, Mitchell provides a “taxonomy” of complexity, distinguishing between constitutive, dynamic, and evolved complexity. While constitutional complexity applies to systems that are composed of different kinds of parts, such as a multicellular organism, dynamic complexity is related to mathematical features of descriptions of the processes occurring in certain systems, such as non-linearity or self-organization. Evolved complexity, finally, refers to diversity that is due to the contingency and randomness involved in evolutionary processes. However, it is clear that Mitchell thinks of these different types of complexity as related in some way given that “complexity” in an unqualified sense plays a central role in some of her general arguments. This raises the question whether it is possible to say something non-trivial about the common core underlying these different types.
The final option, according to which the concept does not play an important theoretical role (or is too fundamental to be of direct relevance), is actually not implausible as far as the usage by biologists is concerned. In most contexts “complexity” is used in a rather casual way and does not figure in actual scientific descriptions or explanations of biological phenomena. There are exceptions, however. In some areas of biology complexity itself becomes a feature of interest or plays a relevant theoretical role. One case is the debate on complexity trends in evolution that was already mentioned in the introduction. Another example comes from parts of ecology, in which complexity is taken to describe the state of an ecosystem and related to other properties, such as diversity or resilience (Parrott 2010). But even if complexity were not of theoretical interest to biologists at all, this would not mean that there is no interest for philosophers to have a clarified idea of biological complexity. In particular, biological complexity does seem to play an important role in philosophical arguments. According to Mitchell,
the complexity of the subjects studied by the various sciences and the limitations of our representations of acquired knowledge jointly entail an integrative, pluralistic model of science. (Mitchell 2003, p. 2)
And Bechtel and Richardson argue
[i]mplicit in our discussion of organization, complexity, and emergence in the previous section is the basis for rejecting ruthless reductionism and for distinguishing mechanistic reduction from ruthless reduction or any other account that construes lower levels as the source of all explanation. (Bechtel and Richardson 2010, p. xxxvii)
It should be noted that similar “metascientific” arguments from complexity are also sometimes put forward by scientists themselves. For example, some systems biologists argue that reductionist approaches are misguided because of the complexity of biological systems:
Biological systems are extremely complex and have emergent properties that cannot be explained, or even predicted, by studying their individual parts. The reductionist approach (...) underestimates this complexity and therefore has an increasingly detrimental influence on many areas of biomedical research. (Van Regenmortel 2004, p. 1016)
Given that complexity figures as an essential ingredient in important metascientific arguments, it does seem to be quite relevant what complexity means in biological contexts, and it is unlikely that an appeal to an intuitive notion of complexity will be sufficient to properly assess the merits of these kinds of arguments.
In summary, I have suggested that, even though omnipresent in the work of biologists and philosophers of biology, the meaning of “complexity” is rarely made precise in biological contexts. Furthermore, I have argued that there are good reasons to insist on a further clarification of the term. In particular, this is important because there are arguments about science, advanced both by scientists and philosophers, in which a notion of biological complexity figures as an essential ingredient. If complexity does not have a clear meaning, or if there are conflicting ideas about what complexity actually consists in, then a proper assessment of these kinds of arguments will not be possible.
3 Clarifying complexity
In this section I introduce several major distinctions that suggest that “complexity” has multiple and very different meanings. In the end, however, I argue that it is possible and productive to consider these distinctions from a unified perspective.
3.1 Ontic and epistemic complexity
As a first step of clarification, it is useful to notice that there are generally two broad senses in which the term “complexity” is applied in the scientific realm. First, complexity is studied as an interesting property of particular systems. Scientists investigate the ways in which systems are complex or exhibit complex behavior, and they try to explain how complexity as a feature of the world can evolve or emerge under certain circumstances. In line with Hans-Jörg Rheinberger, I will refer to this sense as ontic complexity (Rheinberger 1997; see also Kaiser et al. 2014). On the other hand, scientists speak of “complex problems”, just as we do in everyday life, to indicate that they are particularly difficult. In this sense “complexity” does not directly refer to the object under study itself, but to a task, usually cognitive, that relates to the understanding, prediction or control of its behavior. For this sense I will use the term “epistemic complexity”.Footnote 1
The distinction between the complexity of a system and the complexity of a problem will perhaps seem obvious. However, when talking about biological complexity both scientists and philosophers often tacitly shift between these two meanings of complexity. The following quote from Warren Weaver’s 1948 article Science and Complexity may serve as an example:
Living things (...) present situations in which a half-dozen, or even several dozen quantities are all varying simultaneously, and in subtly interconnected ways. Often they present situations in which the essentially important quantities are either non-quantitative, or have at any rate eluded identification or measurement up to the moment. Thus biological and medical problems often involve the consideration of a most complexly organized whole. (Weaver 1948, p. 536, emphasis added)
Here complexity refers, on the one hand, to the “subtly interconnected ways” in which a system is organized as a “most complexly organized whole” and, therefore, to intrinsic features of the system. On the other hand, Weaver invokes the fact that our information about the system is limited, because some quantities have “eluded identification or measurement”, which suggests complexity in an epistemic sense.
While it seems very plausible that those systems that are ontically complex are also the ones that are hard to study, the link is perhaps not as obvious as it may seem at first, and it does not justify a conflation of the two concepts. If complexity is understood as an intrinsic property of a system, it should not depend on the state of knowledge of the investigator and the currently available tools of analysis. By contrast, if complexity is understood in an epistemic sense, we must precisely take into account the investigator’s particular cognitive limitations and her access to information about the system. As I will show in the following, there is a clear link between the two concepts because ontic complexity can be understood in terms of the complexity of describing particular aspects of a system, and thus in terms of an epistemic task. However, the link is non-trivial because many epistemic tasks referred to as complex go beyond the merely descriptive, for example by relating different descriptions with the aim of providing a scientific explanation.
3.2 Emergent and mechanical complexity
We have already seen above that there is a certain tension between different intuitions about complexity, even if the concept is clearly understood as a property of biological systems, and thus used in an ontic sense. One intuition is that complexity is basically a measure of the complicatedness of a system, which suggests a close connection with complexity in an epistemic sense (because complicated systems are difficult to understand). On the other hand, complexity is often associated with order and organized structure, which suggests that the link between ontic and epistemic complexity is not as straightforward.
A notion of complexity that is related to order and structure seems to be the main focus of what is often called “complexity science”. In this context, complexity is typically understood as a property exhibited by certain classes of dynamical systems. These systems are characterized by features like chaos, nonlinearity, or self-organization. Starting in the 1960s and 1970s, in the course of what some have referred to as the “complex systems revolution”, systems with such features came to be widely studied by mathematicians, physicists and other theoretical scientists (Hooker 2011).
Ladyman et al. (2013), and more recently Ladyman and Wiesner (2020), have attempted to provide a tentative definition of complexity which is very much in the spirit of complexity science. According to their analysis
[a] complex system is an ensemble of many elements which are interacting in a disordered way, resulting in robust organization and memory. (Ladyman et al. 2013, p. 57)
One often invoked paradigm for such systems is a flock of birds, a system which consists of individual units that locally interact more or less randomly, but that give rise to robust and coherent patterns at a higher level without requiring any kind of central control or pre-determined program. Proponents of this perspective often conceive of complexity as an emergent property. I will therefore refer to it as emergent complexity.Footnote 2
While this is certainly one way in which the term “complexity” is used, and while we will later see that there are scientists who think of living systems as complex and self-organized in precisely this way, there seem to be aspects of biological complexity that are not captured by this perspective. Consider the following quote from a textbook in molecular biology:
The increased complexity of eukaryotic replication machinery probably reflects more elaborate controls. For example, the orderly maintenance of different cell types and tissues in animals and plants requires that DNA replication be tightly regulated. (Alberts et al. 2015, p. 254)
This doesn’t sound as if complexity simply “emerged” from lower level disorder. The ways in which the components of a eukaryotic cell interact appear to be extremely specific and have been shaped by a long evolutionary process. Rather than originating in lower level randomness, the complexity of the system seems to be related to the fact that its parts are tightly controlled and cooperate in a highly orderly fashion, much like a mechanical system in which each component plays a very specific role. The paradigm of such a system is of course the mechanical clock. If we take Ladyman et al.’s definition seriously, then a clock is not a complex system at all because it does not consist of elements interacting in a disordered way, and, probably, because a clock is not robust in the relevant sense. This suggests that there is a second important sense in which “complexity” is used by biologists, which I will refer to as mechanical complexity.Footnote 3
The two senses of complexity that I have just delineated should not be thought of as referring to radically different kinds of objects, but rather as two extremes of a spectrum. In fact, if asked directly, most biologists would probably agree that regarding their complexity biological systems are situated somewhere between mechanical clocks and flocks of birds.
The difference between the two kinds of complexity bears some similarity to the distinction made by Warren Weaver between “organized” and “disorganized complexity”. Weaver describes a problem of disorganized complexity in the following way:
It is a problem in which the number of variables is very large, and one in which each of the many variables has a behavior which is individually erratic, or perhaps totally unknown. However, in spite of this helter-skelter, or unknown, behavior of all the individual variables, the system as a whole possesses certain orderly and analyzable average properties. (Weaver 1948, p. 538)
The paradigmatic illustration for disorganized complexity in Weaver’s sense is the statistical description of a gas. While it is completely impossible to determine the trajectories of the individual molecules constituting the gas, one can describe higher-level regularities with the help of statistical methods, averaging over the behavior of the molecules. Problems of organized complexity, by contrast, involve “dealing simultaneously with a sizable number of factors which are interrelated into an organic whole” (Weaver 1948, p. 539). According to Weaver, these kinds of problems are predominantly found in biology, but also arise in the psychological, economic, and political sciences.
We have to note, however, that Weaver’s idea of disorganized complexity does not exactly match Ladyman et al.’s definition because Weaver also includes systems in which there is no organization at all. In Ladyman et al.’s view, by contrast, complexity has to be found somewhere between perfect order and complete disorder.
In Sect. 4 I will suggest a way in which all these different ideas can be taken into account within a single framework. Before getting there, however, one further issue has to be addressed.
3.3 Complexity of mechanism and complexity of behavior
When observing how biologists use the term “complexity”, one notices a further distinction that has not yet received much attention in philosophical discussions. Compare the following statements:
Although they are single cells, protozoa can be as intricate, as versatile, and as complex in their behavior as many multicellular organisms. (Alberts et al. 2015, p. 30).
Although bacteria also have cell memory mechanisms, the complexity of the memory circuits in higher eukaryotes is unparalleled. (Alberts et al. 2015, p. 206)
In the first case, “complexity” refers to the behavior of a system. The attribution of complexity seems to derive from the fact that some capacity or range of capacities of the system is in some sense impressive. In the second case, “complexity” refers to a mechanism. Here, the attribution of complexity seems to express the idea that the structure or the organization of the system is complicated. Thus, it is natural in the biological context to conceptually distinguish between the complexity of behaviors and the complexity of mechanisms.Footnote 4 While this distinction bears some similarity to the distinction between ontic and epistemic complexity, and also to the distinction between mechanical and emergent complexity, it introduces a new aspect because it can be understood as relating different meanings of complexity respectively to the explanans and to the explanandum in biological explanations. In many areas of biology the explanans is provided by the description of a mechanism, while the explanandum is a phenomenon or behavior of interest (Wimsatt 1974; Machamer et al. 2000).
Complexity of behavior is related to the capacities of a biological system, for example to the number of functions it can perform, to its adaptive response to varying contexts, or to its robustness in the face of perturbations. Complexity of mechanism, by contrast, is more straightforwardly related to how complicated a system is, for example to the number of components, to the complexity of interactions (e.g. linear vs non-linear), or to its topological structure. A very clear example is provided by the central nervous system, which is often called complex with reference to its behavior, i.e. because of the many different tasks it controls or is involved in, and because of its capacity for adaptive behavior and learning. On the other hand, it is called complex with reference to its structure, i.e. the architecture of different parts and the unimaginably large number of neurons and their patterns of connection.
A priori these are two independent notions, but biologists of course expect that the complexity of a behavior and the complexity of the underlying mechanism will more or less match in degree. However, cases where these kinds of expectations are confounded, i.e. when seemingly complex behaviors are the result of surprisingly simple mechanisms, or when the mechanism underlying a simple behavior appears unnecessarily complex, can be particularly instructive. Instances of the former are found for example in developmental biology, where the mechanisms underlying complex morphological patterns have often been found to be simpler than expected (e.g. von Dassow et al. 2000; Lewis 2003). Interestingly, these are typically cases where intuition fails and mathematical methods have to be employed to explain the phenomenon in terms of the underlying mechanism. This shows that epistemic complexity, i.e. the complexity of the explanatory task, is not related in a straightforward way to the complexity of the system.
To summarize this section, I have discussed three distinctions that should be taken into account when looking for a clarified picture of biological complexity. Ontic and epistemic complexity apply to different categories: ontic complexity directly refers to the systems studied by scientists, whereas epistemic complexity refers to a scientific task or problem. Mechanical and emergent complexity are two different and somewhat conflicting ways in which the notion of ontic complexity can be further spelled out. Mechanical complexity captures the intuition that a system is complex if its components interact in very specific and tightly controlled ways, whereas emergent complexity is the idea that a system is complex if it exhibits some degree of structure or organization that arises from underlying disorder. Finally, the distinction between complexity of mechanism and the complexity of behavior was introduced to take into account the fact that biologists, even when talking about the same system, apply the notion of complexity differently depending on whether they use it with reference to the explanans or the explanandum. It is important to note that in my account mechanical complexity and complexity of mechanism are very different notions. In my usage, “mechanism” or “mechanistic” refers primarily to the logical role of being the explanans in the (causal) scientific explanation of a phenomenon, while “mechanical” refers to a particular idea of the structure and organization of a system.
4 A unified framework for biological complexity
What I have called emergent and mechanical complexity seem to be quite different, even diametrically opposed notions. From the point of view of the emergent complexity conception, mechanical complexity is not complexity at all since the order found in a mechanically complex system does not arise from disorder. Conversely, from the point of view of mechanical complexity, a system like a flock of birds is not complex because its behavior does not seem to depend on the fine details of the structure of the system. The distinction between complexity of mechanism and complexity of behavior adds to the heterogeneity of the resulting picture.
In this section I propose a more encompassing perspective that aims at capturing these different aspects based on the technical notion of effective complexity, introduced by Murray Gell-Mann and Seth Lloyd (Gell-Mann and Lloyd 1996; Gell-Mann 2002). The basic idea underlying my framework is that biological complexity should be understood as a quantitative relationship between the effective complexity of the behavior of a system and the effective complexity of the underlying mechanism. This allows me to understand emergent complexity and mechanical complexity as different regimes of this relationship. At the same time it illuminates the way in which biological complexity is necessarily relative and cannot be attributed to systems in an unqualified manner.
4.1 Effective complexity
As we have seen, it is a common idea that complexity is not a simple concept itself, and that there is a plurality of views on the actual nature of complexity. Independently of this fact, it might nevertheless be possible to find a measure of complexity that enables comparisons of different systems. This is not at all uncommon in science. For example, measures of temperature were accepted long before there was any agreement on the nature of the underlying phenomenon.
One quickly recognizes, however, that there are not only different perspectives about what complexity is, but also that there exist numerous different measures of complexity that are applied in different contexts (Lloyd 2001). Many complexity measures are in some way based on numerosity, or the number of relevant components, processes, interactions etc. of a system (cf. Ladyman and Wiesner 2020). Similarly, in the context of statistical and mathematical modeling, complexity is often related to the number of variables or of free parameters of a model. In biological contexts, the complexity of organisms is often assessed based on the number of different genes or of cell types. Other measures are based on variance. For example, McShea (2005) proposes to compare the degree of internal variance between different organisms in order to investigate possible complexity trends in evolution. The fact that there are different measures of complexity in different contexts does not necessarily imply a multitude of underlying concepts. These different measures might be understood as proxies that in different ways provide information about the actual underlying complexity of a system. A common trait of all these measures is that they directly relate the complexity of a system to some property of a description of that system.
A generalized and formal version of this kind of measure can be found in the concept of algorithmic complexity (Kolmogorov 1963), or algorithmic information content, which is defined roughly as the length of the shortest set of instructions that can reproduce a given string of letters or numbers. One may thus feel inclined to say that a system is more complex than another if its description has higher algorithmic complexity. However, a tension with intuitive ideas about complexity arises here because maximal algorithmic complexity is assigned to a fully random string. If we were to directly apply the idea of algorithmic complexity to the description of systems, then a completely disordered system, such as a gas, would have to be considered maximally complex because we need to describe the motion of each particle independently and have no way of compressing that information. This suggests that a reasonable general measure for the complexity of systems must take into account in some way the degree to which the system is structured or organized.
Gell-Mann and Lloyd (1996) proposed a measure of “effective complexity” in order to capture this aspect. Gell-Mann explains the idea as follows:
It would take a great many different concepts to cover all our intuitive notions of what is meant by complexity (and its opposite, simplicity), but the concept that agrees best with the meaning in ordinary conversation and in most scientific discourse is effective complexity (EC). Roughly, the EC of an entity is the length of a very concise description of its regularities. (Gell-Mann 2002, pp. 13–14)
Thus, complexity is clearly related to algorithmic information content because Gell-Mann and Lloyd understand “very concise description” exactly in terms of the shortest program generating the description of an entity. But importantly, the relevant description is restricted to certain features of the entity. More specifically, they argue that the total algorithmic information can be decomposed into two terms that can be understood as describing the random and the regular features of an entity, respectively. In order to achieve this decomposition, the entity of interest must be embedded within an ensemble of comparable entities. These entities share certain features but may be different in various other respects. Effective complexity is then defined as the algorithmic information content of the ensemble. The choice of ensemble therefore determines what the regular aspects of an entity are. The remaining part of the total algorithmic information content of the entity is interpreted as a description of its non-regular features. It can be understood as an entropy term because it measures our ignorance about the entity if we would only know that it belongs to the ensemble.Footnote 5 Intuitively, the larger this term, the more the members of the ensemble can vary among each other.
The common idea behind both algorithmic complexity and effective complexity is that they define complexity in terms of the properties of descriptions, i.e. of linguistic objects that can in principle be represented by a sequence of symbols or string. Thus, in order to illustrate the basic idea, let us consider the following two strings of binary numbers:
While the first string looks essentially random, the second exhibits a very simple pattern. In line with this, the algorithmic complexity of \(s_2\) is smaller than that of \(s_1\). This is because one can generate \(s_2\) with the instruction to repeat [0, 1] ten times, and thus by a relatively short program. By contrast, there does not seem to be a way to reduce the description of \(s_1\) and thus an instruction to write out the string needs to contain some representation of the whole string itself.
In many contexts, notably in biology, we would not ascribe maximum complexity to completely random configurations, but rather think of objects as complex if they exhibit some degree of structure or organization. Applied to our toy example, what we want to find is a measure that assigns higher complexity to \(s_2\) than to \(s_1\). We can imagine, for example, that the two strings represent the outputs of two differently oriented telescopes, \(T_1\) and \(T_2\), that measure electromagnetic radiation coming from space. Now let us assume that we repeat the measurement for both telescopes several times and that for \(T_1\) we always get a different but random looking string, and that for \(T_2\) we always get a string with alternating 0s and 1s. We would then be inclined to say that what we measure for \(T_1\) is probably noise, and that what we measure for \(T_2\) is an actual signal. In line with this scenario, the idea of effective complexity is to think of the particular object under study as a typical member of a larger ensemble and to consider the information that is needed to describe this ensemble as a measure of its complexity. Thus, the string \(s_1\) has low complexity if we think of it as a typical member of the ensemble of all binary strings of length 10 because this ensemble simply has no specific properties that need to be described. And \(s_2\) has higher complexity than \(s_1\) if we consider it as a representative of the ensemble of binary strings of length 10 with alternating 0s and 1s because we have to provide additional information to define this ensemble.
From this perspective complexity is not an intrinsic property of the entity, but depends on the particular choice of ensemble. To see this, consider an alternative scenario where repeated measurements of \(T_1\) always result in exactly the same string \(s_1\). In this case we would likely not think of it as noise but rather as a complex signal. Effective complexity makes sense of this: the relevant ensemble has changed and does not consist of all strings but only of \(s_1\) itself. Therefore, the effective complexity of \(s_1\) is now higher than that of \(s_2\) because the description of this ensemble is considerably longer than the description of the ensemble of \(s_2\).
Effective complexity matches the intuition that complexity is somehow related to complicatedness (in this context the difficulty of describing a string), but only if this complicatedness is relevant according to some external criterion which determines the regular and random features of an entity, respectively. Note that there is some ambiguity about the meaning of “regularity” in this context. Regularity can refer to a pattern within a given entity, such as the alternating occurrences of 0 and 1 in \(s_2\). But it can also refer to a seemingly random pattern that recurs in different entities (such as the irregular string \(s_1\) in the second scenario). Regularity in the former sense is related to simplicity because it allows us to compress the description of a system. Regularity in the latter sense is related to complexity because it suggests the importance of features that need to be taken into account in a description.
Taken together, effective complexity makes sense of the two seemingly conflicting intuitions that complexity depends to some extent on regularity or order, but is at the same time related to complicatedness.
4.2 The relativity of complexity in biology
How can this idea be applied to real systems, and in particular to the context of biology? While the actual definition of effective complexity in terms of a decomposition of algorithmic information content is quite technical, the intuitive idea behind it actually comes close to what biologists are usually interested in when describing phenomena or their underlying mechanisms. In most cases, biologists do not aim to describe a particular entity in all its idiosyncratic detail, but rather to describe features common to a particular class of entities (whether those objects are macromolecules, cells, organisms, or ecosystems). Thus biology, as many other scientific disciplines, can be seen as engaged in the study of ensembles of entities that share a set of relevant features despite some random variation between them. Effective complexity matches these practices well because it relates complexity to the description of these relevant features. At the same time it makes sense of the seemingly paradoxical fact that complexity is both somehow related to complicatedness and to order. Biologists think of a multicellular organism as more complex than a random bag of cells, even though it seems more difficult to describe the latter at the same level of detail. In analogy with the random string \(s_1\) in Sect. 4.1, a description of the random bag of cells would require explicit specification of the location of each cell. An organism, by contrast, can be described in a more economical way because it is organized into spatially separated tissues and organs which contain certain cell types and not others. Nevertheless, the random bag of cells is considered less complex because it is implicitly treated as a system that has no particular configuration (even though every single bag of cells does have a particular configuration), and thus as a member of a large ensemble of entities that do not share any interesting features.
An important consideration to be taken into account, however, is that many formal measures of complexity are defined with respect to idealized model systems. In particular, they presume a canonical description of a system which uniquely determines its complexity. For real-world systems, by contrast, complexity is not uniquely defined even given a fixed complexity measure because it also depends on the choice of representation. One may represent a biological organism using just its weight and the position of its center of mass, or one may describe all its components in detail down to the molecular level. More generally, there is not necessarily one privileged decomposition of a system into parts, nor even a unique way of determining what the system’s behavior is. As Stuart Kauffman observes,
not only are multiple views about what a system is doing possible, but also any system may be decomposed into parts in indefinitely many ways, and for any such part, it too can be seen as doing indefinitely many things. (Kauffman 1976, p. 259)
William Wimsatt, drawing on Kauffman’s ideas, argues that, as long as we do not have one exhaustive and unifying theory, each theoretical perspective taken by itself can only give an impoverished view of the real objects (Wimsatt 1972, reprinted in Wimsatt 2007, Chapter 9). In particular, it seems that when we call a system “complex” from a given theoretical perspective, we can in effect only judge the complexity of our particular representation. This suggests that, in order to get an idea of the “actual” complexity of a system, we must also consider how different theoretical perspectives relate to each other. In this regard, Wimsatt offers two concepts—descriptive complexity and interactional complexity—that may be seen as proxies for the possibly inaccessible “actual” complexity of a system. A system is descriptively complex to the extent that different theoretical perspectives pick out decompositions into parts that do not spatially coincide. Scientists belonging to different biological disciplines decompose an organism, like a fruit fly, differently into parts, e.g. according to cell types, developmental fields, or physiological systems. In more descriptively simple systems the decompositions according to different perspectives will tend to coincide more.
In order to understand Wimsatt’s slightly more complicated concept of interactional complexity, we have to consider the system in a state space representation, that is, in a representation that describes the behavior of variables characterizing the system over time. Each theoretical perspective picks out different properties of a system and therefore works with a different set of variables. Depending on the desired level of predictive accuracy, one can neglect causal links below a certain threshold of interaction strength and thereby obtain a decomposition into subsystems with strong internal bonds. A system is interactionally complex if many of these subsystems partly fall into different perspectives. One will neglect important causal factors and not be able to predict its behavior with precision, unless one considers it from more than one perspective.
Wimsatt’s analysis thus highlights a further important aspect of relativity: when assessing the complexity of a system, not only must different theoretical perspectives be taken into account, but also the desired level of precision. A system that seems to behave in a very simple fashion when represented in a relatively crude way, such as a mass of water flowing through a tube, appears extremely complex as soon as we aim for a more precise description in which factors like viscosity and turbulence cannot be neglected anymore. Thus, each theoretical perspective seems to give us only a rough approximation of the “actual” complexity of a system and at best a lower bound.
Does this mean that any non-relative account of ontic complexity is out of reach? Wimsatt suggests that it is difficult, or even pointless, to strive for an account of the complexity of a system in an absolute sense. But maybe such an absolute concept is not what is needed anyway in order to understand the role of the concept of complexity in biology. If we take into account how and when biologists usually apply the concept, it is not clear that a non-relative concept of complexity is even relevant. And the distinction introduced in Sect. 3.3, between the complexity of mechanisms and the complexity of behaviors, may in fact prove to be a solution to the problem of relativity rather than an additional complication. As we have seen, biologists typically apply the concept of complexity not simply to systems in an unqualified sense. Thus, they do not ask “how complex is a fruit fly?”, but instead ask, for example, about the complexity of the network underlying segmentation of the fruit fly embryo. Similarly, when it comes to comparisons between different kinds of organisms, say, about the differences between humans and other animals, what is at issue is not a difference in complexity per se, but rather a difference in complexity with respect to a particular set of biological capacities.
Thus, in biological contexts complexity is rarely unconditionally applied to systems, but either with reference to a specific behavior of a system or to a mechanism underlying a specific behavior. The idea of complexity as a property that is unconditionally applied to systems may make sense from the perspective of complexity science with its focus on formally specified systems, but it does not fit the situation of biology.
The focus on mechanism and behavior allows us to elaborate on the application of the concept of effective complexity in biology. As we have seen, embedding a system in an ensemble effectively means to decide which are the relevant regularities of the system. For the case of mechanisms this kind of relevance relation can in principle be derived from the norms of mechanistic explanation. Thus, the relevant ensemble of a mechanism underlying some behavior can be defined in terms of the regularities that would need to be included in a description that counts as a successful explanation of the behavior.Footnote 6 A similar strategy for accounts of biological behaviors appears more problematic, however, because it seems that how these are described depends to a large degree on the particular interests of the investigator. Does this threaten any objective account of biological complexity after all? We can save some degree of objectivity if we take into account the hierarchical structure of explanations in biology. The behaviors that are the explananda in mechanistic explanations in biology are very often embedded in a functional context themselves and figure as parts of the explanantia of yet other behaviors (cf. Craver 2007). For example, the behavior of the heart is explained by the pumping activity of the atria and ventricles, and the behavior of these components is in turn explained with reference to the activity of components at an even lower level. This suggests that descriptions of mechanisms and of behaviors are closely related in biology.
McAllister (2003) argues that effective complexity is not a good measure of information content because it is not uniquely defined and relative to the cognitive and practical interests of investigators. It is not clear to what extent this is really a problem for Gell-Mann because he admits that his concept is relative in several regards. But it certainly does not pose a problem for my conception of biological complexity because the relevant features, or regularities, on which the application of the concept of effective complexity depends are not arbitrarily determined by the observer, but are constrained by the larger research context and the overarching aim of providing scientific explanations.
4.3 Mechanical and emergent complexity reconsidered
Given this rough sketch of how the complexity of mechanisms and behaviors can both be understood using the idea of effective complexity, we can now turn to the question if we can make sense of the seemingly very different ideas of emergent and mechanical complexity using this concept as well. I propose that we can consider both concepts as derivative of the idea of effective complexity if we think of them as different ways of relating the complexity of a behavior and the complexity of the corresponding mechanism. Briefly, mechanical complexity describes instances where the effective complexity of the behavior more or less matches the complexity of the mechanism, whereas emergent complexity describes instances where the effective complexity of the behavior is high compared to the complexity of the mechanism.
Recall that the idea of mechanical complexity was that of a system in which the activity of each part is tightly controlled such that the behavior of the system results from the orderly interplay of its components. A satisfactory description of the mechanism underlying this behavior must be relatively detailed. Put differently, the relevant ensemble is relatively small because many possible configurations of the same parts must be excluded. As a consequence, high mechanical complexity is directly related to high effective complexity of the mechanism. At the same time we would probably think of such systems as complex only to the extent that the complexity of the mechanism translates into something that the system is capable of doing. Thus, in mechanically complex systems the complexity of mechanism and the complexity of behavior are roughly of the same order. Given the evolutionary history of biological systems and the contingent and random aspects of evolutionary processes, it is plausible to find many instances in which the complexity of a mechanism seems out of proportion with the the complexity of the corresponding behavior. Elliot Sober cites the example of the surprisingly long loop in the tubing connecting the testes to the penis in mammals as an example of a clear violation of “minimality” (Sober 2015, p. 153). The loop is a result of the particular evolutionary history and does not apparently contribute to the functionality of the structure. Singh and Gupta (2020) have recently proposed the idea of “unnecessary complexity” to describe deviations from “the minimum number of gene-gene interactions and minimum biochemical path lengths necessary for a given molecular function, trait or an organism” (Singh and Gupta 2020, p. 3). More generally, there are usually multiple ways to solve an adaptive problem, and it is widely accepted that evolution does not necessarily produce the optimal solution (e.g. Parker and Smith 1990), let alone the minimally complex solution. Thus, I propose to speak of mechanical complexity when the complexity of the mechanism is similar to the complexity of the behavior, and of unnecessary complexity when the complexity of the mechanism is considerably higher than the complexity of the behavior.
In instances of emergent complexity the relationship between the complexity of the mechanism and the complexity of the behavior is quite different. Here, we have relative simplicity at the level of the mechanism and relative complexity at the level of the behavior. Consider again the example of the flock of birds. Organized and orderly behavior at the level of the flock is thought to “emerge” from disorder and relatively simple interactions at the level of the individual birds. Disorder in this case means that the initial exact configuration of birds does not matter, and that one needs relatively little amount of detail to describe the setup of the system. The relevant ensemble therefore consists of a large number of possible “bird configurations”. The idea of low complexity in mechanism and high complexity in behavior also matches the standard theoretical examples of complexity science, such as the logistic map or the Lorenz system.Footnote 7 In both cases there is a relatively simple description of the system in terms of the generating equations (corresponding to the mechanism), but an unexpected complexity in the behavior exhibited by the system. Note that low complexity of a mechanism does not imply that it is easy to understand how the mechanism brings about the behavior. Explaining complex behavior in terms of a simple mechanism is typically a task of high epistemic complexity and often requires sophisticated mathematical tools. By contrast, explaining how the many components of a mechanical clock bring about its time-keeping behavior is surely complicated as well, but seems rather straightforward by comparison. This is presumably another reason why many people are inclined to consider emergent complexity as the only relevant kind of complexity.
In Fig. 1 three possible ways of combining the complexity of mechanism and the complexity of behavior are represented in a schematic diagram. This representation also suggests that there is not a clear separation between the different kinds of complexity, but rather a continuous transition. A quantitative measure for the complexity c(S) of a system S that captures both emergent complexity and mechanical complexity may tentatively be provided by the following expression:
Here, \(c_B\) stands for the complexity of the behavior of S, and \(c_M\) stands for the complexity of the mechanism underlying S’s behavior. First of all, the equation expresses the idea that overall complexity directly increases with the complexity of behavior. The factor \(\kappa \) takes into account the relationship between the complexity of mechanism and the complexity of behavior. We can think of it as the “emergence factor” in the following sense: Mechanical complexity corresponds to systems for which \(\kappa \approx 1\), and thus complexity is directly proportional to the complexity of its behavior. Emergent complexity is present when \(\kappa \gg 1\), and the case of \(\kappa \ll 1\) corresponds to unnecessary complexity.Footnote 8
This relationship captures the idea that a system like the flock of birds is considered complex not because it exhibits behavior that is particularly complex in absolute terms, but because its behavior is complex relative to the complexity of the underlying mechanism. Furthermore, it is consistent with Ladyman et al.’s sense of complexity emerging from underlying disorder because \(c_M\) is inversely related to the entropy or degree of disorder, which directly follows from Gell-Mann (2002)’s analysis of total algorithmic complexity as the sum of effective complexity and entropy. It also captures in a rather straightforward sense the worn-out but rarely clarified idea that emergence is present when the whole (i.e. the system’s capacities or behavior) is greater than the sum of its parts (i.e. the underlying mechanism). Finally, the equation also shows in what sense mechanical and emergent complexity can be understood as lying on opposite ends of a spectrum because given a particular explanandum phenomenon (i.e. \(c_B(S)\) is held fixed), a proposed mechanism exhibits mechanical complexity if \(c_M(S)\) is high and emergent complexity if \(c_M(S)\) is low.
I am not hereby suggesting that this kind of “pseudo-quantitative” measure is directly useful in scientific practice. But I think that it provides a productive way of thinking about biological complexity. Even though it may not get at the “essence” of complexity and does not tell us anything about whether there is an underlying biological or physical property that unites all complex systems, it seems to capture our intuitions well when we compare the complexity of different systems. In particular, it is applicable to both mechanical and emergent complexity, and it is directly related to the basic idea of effective complexity as whatever it is that makes our description of the relevant features of an entity long. Thus, there is not necessarily one underlying intrinsic feature that unifies all systems that we would like to call “complex”. Instead, numerosity, internal variance, or interconnectedness, but also dynamic features such as non-linearity and chaos can all contribute to the complexity of a system in this sense. The concept also partially illuminates the relation between ontic and epistemic complexity because it relates complexity to the degree to which we can compress the description of an entity. Badii and Politi who aim at a comprehensive discussion of the ways in which “complexity manifests itself in nature” (Badii and Politi 1999, p. xi) are drawn to a very similar conclusion:
[T]he concept of complexity is closely related to that of understanding, in so far as the latter is based upon the accuracy of model descriptions of the system obtained using a condensed information about it. Hence, a “theory of complexity” could be viewed as a theory of modeling, encompassing various reduction schemes (elimination or aggregation of variables, separation of weak from strong couplings, averaging over subsystems), evaluating their efficiency and, possibly, suggesting novel representations of natural phenomena...[A] system is not complex by some abstract criterion but because it is intrinsically hard to model, no matter which mathematical means are used. (Badii and Politi 1999, p. 6, emphasis added)
In line with this view, complexity can only be assessed with regard to our representations of mechanism or behavior. A system is complex to the extent that it resists ‘condensing’ the amount of information that is needed for describing it. The last part of the quote suggests that this conception, even though subject-dependent in an obvious sense, is not necessarily ‘subjective’ in the sense of depending on the contingent capacities of a particular cognitive agent.
Consider again Warren Weaver’s distinction between ‘disorganized’ and ‘organized’ complexity. The former applies to systems in which the number of variables is very large, but each variable individually shows “helter-skelter” behavior like the molecules in a gas (Weaver 1948, p. 538). When dealing with such systems, one can often apply statistical methods in order to find a compact description in terms of the average behavior of the components. What Weaver is effectively suggesting, therefore, is that disorganized complexity in many cases is just simplicity in disguise. Sometimes, as in the case of statistics, the introduction of a new analytical method can reveal that a system is not as complex as had previously been thought. A similar view can be found in Herbert Simon’s reasoning about complexity. He argues that,
[h]ow complex or simple a structure is depends critically upon the way in which we describe it. Most of the complex structures found in the world are enormously redundant, and we can use this redundancy to simplify their description. But to use it, to achieve the simplification, we must find the right representation. (Simon 1962, p. 481)
This at first seems paradoxical: simplicity is found in complex structures. And in fact, many features that are considered hallmarks of complexity, such as hierarchical organization or modularity, are actually regularities that allow for a simplified description of a system. The idea of effective complexity makes sense of this paradox: if one has to describe a particular entity, the description will be shorter if the entity contains some regularities. By contrast, if one has to describe an ensemble of entities, the description will be shorter if the entities do not share any regularities.
A second aspect of the paradoxical relationship between simplicity and complexity is illuminated by the idea of emergent complexity. I have argued that complexity is often associated with systems that exhibit complex behavior, even though the underlying mechanism appears simple. The relationship between ontic and epistemic complexity, however, is non-trivial because the scientific task of understanding does not consist only in describing mechanism and behavior, but in relating the two descriptions.
5 Illustrating the framework
I would like to illustrate the framework developed in the previous sections with concrete examples from research in biology. In particular, I will discuss different attempts to understand intracellular organization in terms of pathways and in terms of networks. At first glance these attempts seem to imply rather different ideas about biological complexity. While the differences partly concern quantitative aspects of complexity, i.e., the degree of complexity of the system under study, other aspects may best be understood as disagreements about the kind of complexity found in biological systems. But, as highlighted in Sect. 3.2, I suggest that there is a spectrum of complexity between the two extremes of what I have called mechanical and emergent complexity, and so different views on intracellular organization can be located along this spectrum.
Given that I talk about different “perspectives” in the following, one may think that this section mainly discusses epistemic aspects of complexity. However, these perspectives refer to different ways in which scientists conceive of the ontic complexity of biological systems. These different ideas of ontic complexity in turn affect the epistemic complexity of their research problems.
5.1 The pathway perspective
Many biological processes at the molecular level are conceptualized in terms of pathways. Biochemistry studies metabolic pathways that connect a series of chemical reactions and convert one or more reactants into a product. In molecular biology the idea of a molecular pathway is closely linked to the metaphor of biological processes as processes of information transfer or of signaling. The basic idea is that the cell receives an external signal, typically in the form of a molecule binding to a receptor expressed on the cell surface, which in turn transduces this signal via a cascade of steps into the nucleus, where it initiates a specific cellular reaction by switching on or switching off a set of target genes.Footnote 9 A major incentive for the study of such signaling pathways comes from the field of oncology and is based on the idea that cancer can be understood as “a disease of aberrant signal processing” (Weinberg 2014, p. 176). In the context of a multicellular organism, cells divide only under special circumstances, for instance during processes of growth or wound healing. Specific molecules, called growth factors, can act as signals to promote cell survival or to induce growth or cell division. Many traits of cancer appear similar to the responses of normal cells to growth factor stimulation. Based on this observation, biologists hypothesized that cancer may arise when some components of these growth factor pathways become permanently activated. They found that many tumor cells express a mutated hyperactive form of a particular protein, called Ras, and this finding provided the first step to uncover one of these pathways. As the cancer biologist Steven Weinberg recalls,
[o]ver a period of a decade (the 1980s), these circuits were slowly pieced together, much like a jigsaw puzzle. The clues came from many sources. The story started with Ras and then moved up and down the signaling cascades until the links in the signaling chains were finally connected. (Weinberg 2014, p. 181)
This strategy seems very common in episodes of mechanism discovery in the life sciences. Discussing the discovery of the mechanism of protein synthesis, Lindley Darden suggests that biologists often apply a strategy of “forward and backward chaining”. That is, they start with known, or hypothesized, components of a mechanism and then attempt to work forward or backward, taking advantage of constraints that the components impose on the possible ways of filling in the gaps. This strategy is well illustrated by the discovery of the Ras-Pathway, which suggests that the idea of a sequential pathway acts as a powerful heuristic in biological research more generally. It reduces epistemic complexity considerably because it is so intimately linked with our general way of conceptualizing causal processes (cf. Bechtel 2011). At the same time, the idea of signaling pathways implies an idea of biological organization as modular in an important sense. Pathways can be studied in relative isolation from each other, and accounts of the activity of different pathways can be integrated to reach a higher level understanding of cellular behavior. The pathway perspective thus also illustrates the applicability of the strategy of decomposition and localization, as described by Bechtel and Richardson (2010). An implication of this perspective is that intracellular organization is actually not as complex as one might expect given the number of relevant components. If different signals are transmitted via specific pathways that are largely isolated from each other, then many potential interactions between components are excluded. And if each component of a pathway interacts with one or at most a few up- and downstream signaling partners, this greatly reduces the number of possible interactions that are expected to be relevant.
The pathway perspective thus suggests a picture of (relatively) low epistemic complexity. Explanations in terms of sequential organization usually do not require sophisticated models and can often be given in informal, qualitative terms. Furthermore, the strategy of functional decomposition allows biologists to black-box lower level processes and to produce tractable explanations in terms of higher level components, which corresponds to a further reduction in epistemic complexity.
Going beyond the epistemic level, the pathway perspective appears as a rather clear instance of mechanical complexity. The behavior of the cell as a whole is envisioned as arising straightforwardly from the contributions of individual and largely independent pathways. Thus, the complexity of the behavior at this level is expected to be of the order of the sum of the complexities of individual pathways. The pathway perspective also implies mechanical complexity if we go down one more level to the explanations of individual pathway behavior. The molecular interactions that constitute a signaling pathway are taken to be extremely specific, and even slight perturbations on individual components of a pathway are expected to result in major problems, as shown in pathologies like cancer. The precise configuration of steps is important and differs from one pathway to another:
This signal transduction biochemistry is organized around a small number of basic principles and a large number of idiosyncratic details. (Weinberg 2014, p. 220)
Thus, while the organization inside a cell may not be as tightly constrained as a clockwork, the pathway perspective suggests that we need to take into account all the idiosyncracic details to explain cellular responses in terms of the behaviors of pathways and pathway behavior in terms of the activities of individual signaling components.
5.2 The network perspective
Many biologists argue that the pathway perspective is overly simplistic and that intracellular organization should be conceptualized in terms of networks instead. In fact, it is not even clear that the biologists involved in the study of signaling pathways themselves subscribe to the idea of biological organization described in the previous section. Weinberg himself admits, for example, that
our depiction of how signals are transmitted is likely to be fundamentally flawed: a signaling input (...) may operate like the plucking of a fiber in one part of a spider web, which results in small reverberations at distant sites throughout the web. (Weinberg 2014, p. 224)
The advent of genomics, that is, of experimental methods that simultaneously measure the abundance of thousands of cellular components, has recently enabled biologists to investigate the structure and organization of molecular networks in detail. Making use of the mathematical concepts of graph theory, such investigations have revealed connection patterns among molecular components that put the picture of neatly separated pathways into question (e.g. Strogatz 2001; Barabási and Oltvai 2004). In particular, it has been shown that many biological networks exhibit so-called scale-free architecture, which means that most nodes have only very few links, while there are a few nodes, called “hubs”, that are highly connected. Scale-free networks are typically robust to a wide range of perturbations because these are likely to affect only the lesser connected nodes, thereby leaving the overall network largely intact. Perturbations affecting hubs, however, are thought to have dramatic consequences for network behavior. In line with this idea, Jørgensen and Linding (2010) argue in a review article called Simplistic pathways or complex networks? that cellular organization is based on networks and that diseases such as cancer should not be understood in terms of perturbed pathways, but rather in terms of “rewired” networks.
At first glance the network perspective seems to provide a very different take on biological complexity. While the pathway perspective restricts the amount of relevant molecular interactions to a manageable number and implies that it is possible to separate functionality within the system, the network perspective allows for widespread interaction. For this reason the basic strategies of structural decomposition and functional localization (Bechtel and Richardson 2010) are not easily applicable to such networks (Green et al. 2018). To reduce epistemic complexity, alternative heuristic strategies are required, such as the strategy of decomposing large networks into smaller “network motifs” (e.g. Alon 2020).
At the level of ontic complexity, however, it is important to see that many varieties of the network perspective mainly involve a quantitative, and not necessarily a qualitative difference in complexity. In particular, many discussions of network organization are framed in terms of the same metaphor of communication that underlies the pathway perspective. Jørgensen and Linding, for example, write:
Cellular signaling networks are information processing systems. They receive, interpret, correct and transmit or propagate input cues to other control layers in the cell, ultimately altering cell behavior or processes (...). Cellular signaling is in many aspects identical to a general communication system (...) where a message (signal) needs to be transferred to a recipient. (Jørgensen and Linding 2010, p. 15)
This description suggests that biological networks share many properties with designed electronic circuits. Each component plays a very specific role that contributes to the overall performance of the signaling system. Non-sequential organization is thought to arise a consequence of the fact that biological systems are exposed to noise because “as error correction becomes important (...) a nonlinear, branched and more complex network is required” (Jørgensen and Linding 2010, p. 15). Thus, the resulting view of complexity is still mechanical in the sense that it relies on the ordered interaction and tight control of the system components. Disorder (“noise”) is not playing a constructive role in this picture, but is rather an undesirable factor that the mechanism needs to be protected from. The main difference to the pathway perspective lies in the fact that the behavior of the cell is not conceptualized as easily decomposable into the sum of the activities of isolated modules. Yet, in line with the pathway perspective, the mechanism (in this case the network as a whole) is thought of as similar in complexity as the cellular behavior to be explained.
This idea of “mechanical network complexity” goes along with a particular perspective on the evolution of biological networks, according to which the details of their organization have to a large degree been determined by natural selection. This perspective is well illustrated by the strategy of network motifs, which consists in the quantitative comparison of the structure of an actual biological network to a class of similar but randomly connected networks in order to discover biologically meaningful patterns (Alon 2020). The underlying assumption of this approach is that among the myriad of possible network topologies that are consistent with the general chemical composition of a cell, the real biological networks are found in a vanishingly small subset. In other words, real networks are topologically special and the ensemble that characterizes their relevant features is relatively small. As we have seen, this means that they have high effective complexity, which is consistent with mechanical complexity if we consider network topology as part of the explanans of cellular behavior.
In summary, while the network perspective suggests a considerable increase in the degree of complexity when compared to the pathway picture, it does not necessarily leave the familiar terrain of mechanical complexity.
5.3 The attractor perspective
A more radically different view of network complexity goes back to the early work of Stuart Kauffman. The basic idea is that many aspects of cellular phenomena, such as differentiation and plasticity, do not depend on the minute details of underlying molecular processes, but can instead be explained in terms of “generic” properties of large networks.
Originally, Kauffman, who began working on these ideas in the 1960s, thought that this was the only expedient strategy for gaining an understanding of certain biological processes given the knowledge available at the time. His key idea was to tentatively interpret the dynamics of living systems as guided by what Weaver called “disorganized complexity”. Just like theoretical physicists had been able to master the apparent complexity of large disordered systems, one might be able to find conceptual tools to reduce the apparent complexity of large biological systems. In line with this strategy, Kauffman referred to his strategy as an “ensemble approach”, adopting this terminology from statistical mechanics.
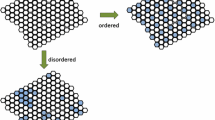
By studying simple network models, Kauffman found some of their properties reminiscent of the behavior of biological cells. The networks he investigated were randomly generated Boolean networks with nodes representing genes that have only two different states: “on” and “off”. The dynamics of these networks unfold as the states of all genes are updated in successive discrete time steps. The state of a gene at a given time point is determined by the states of a set of “input genes” at the preceding time point, which can be interpreted as a set of transcription factors regulating the activity of a common target gene. Since a system defined in this way is deterministic and has a finite number of possible states, it will unavoidably return to a state that it has already previously passed and from then on repeat the same sequence, or cycle. In most networks Kauffman found a surprisingly small number of cycles compared to what would have been combinatorially possible. Moreover, the cycles tended to be relatively short. He interpreted them as corresponding to the different “cell types” that can be exhibited by genetically identical cells. Extrapolating from the relationship between the size of the network and the number of cycles obtained from his simulations, he was able to derive reasonable estimates for the number of different cell types in various species of multicellular organisms (Kauffman 1969).
More recently, Kauffman’s ideas have inspired a research program in systems biology (e.g. Huang et al. 2005). Here cellular networks are interpreted as large dynamical systems with attractor states that correspond to stable cell types. Generalizing Kauffman’s cycles, attractors are understood as generic features of a certain class of dynamical systems. Cellular phenomena such as plasticity, differentiation, or reprogramming are thought to be explainable without the need to invoke the minute details of network structure. In particular, cancer is not explained in terms of a “rewiring” of the network, but as a transition of cells into so-called “cancer attractors” (Huang et al. 2009).
Differently from the other two views presented in this section, the attractor picture may best be understood as an illustration of emergent complexity. It resembles the explanation of the behavior of a flock of birds in several respects. At the level of the explanandum we have a coherent higher level pattern (e.g. the differentiation trajectory of a cell towards its eventual fate) that is maintained even in the face of perturbations. However, this behavior is not explained in terms of tightly controlled and highly specific processes at the lower level, but is thought to follow from relatively unspecific assumptions about the activities and interactions of lower level entities.
In stark contrast with the network motif approach, the attractor perspective relies on the assumption that by investigating typical members of a random collection of networks, one can get an understanding of the particular instances found in nature. This in turn presupposes that, during the process of their evolution, these networks have retained, or acquired, an essentially random structure at the large scale.Footnote 10 Seemingly complex behaviors of biological systems might thus find an explanation that does not require uncovering all of the underlying molecular details:
There is a fundamental ontological assumption underlying [the ensemble] approach, and it is not known if that assumption is true or false. Is it the case that the genetic network in an organism, or a species, or family of species, after 3.8 billion years of natural selection and evolution, is a highly crafted, “one off” design, brilliantly tuned by selection to achieve its functions? Or might it be the case that real genetic regulatory networks are more or less “typical” members of some class, or ensemble, of networks which selection has modified to some degree? In the latter case, we may be able to gain very considerable insight into the structure, logic, and dynamics of gene regulatory networks by examining the typical, or generic properties, of ensemble members. (Kauffman 2004, p. 582)
Thus, in Kauffman’s perspective we see a clear deviation from the idea of mechanical complexity. Complexity of behavior does not depend on the specific details of the underlying mechanism but partly “emerges” from underlying disorganized complexity and thus from a network that is actually relatively simple when compared to the effective complexity presumed in other approaches.
Of course I do not argue that all biologists thinking in terms of pathways are committed to mechanical complexity, and that all proponents of an attractor perspective are committed to emergent complexity. My aim in this section was to present a set of examples that are particularly clear. In practice, however, many cases are expected to lie somewhere on the spectrum between mechanical and emergent complexity (e.g. Bhalla and Iyengar 1999).
6 Conclusion
In this paper I have attempted to clarify the meaning of biological complexity. A better understanding is often precluded by the failure to take into account a number of conceptual distinctions. In particular, I have argued that one needs to distinguish between ontic and epistemic complexity and between the complexity of mechanisms and of phenomena. I have further argued that there are two different versions of ontic complexity, mechanical and emergent complexity, that are relevant in the context of biology and that can be understood as two ways of relating the complexity of mechanism and behavior. Drawing on the concept of “effective complexity” introduced by Gell-Mann and Lloyd, I have proposed a unifying framework that appears to work well in the context of biology and that captures the conflicting intuitions underlying mechanical and emergent complexity.
This framework can be used to understand the differences between alternative perspectives on biological organization. As has been shown, intracellular processes are conceptualized in terms of mechanical complexity by some biologists and in terms of emergent complexity by others. These two perspectives of biological complexity can be directly related to the kinds of ensembles on which explanatory accounts are implicitly based. In Kauffman’s perspective the relevant ensemble is large and contains many apparently random networks. By contrast, in the more “conventional” network approaches, notably in the strategy of network motifs, the relevant ensemble is small in comparison, which is evident from the strategy of explicitly contrasting biologically meaningful networks with random networks. Thus, it turns out that some network approaches share an idea of mechanical complexity with the traditional picture of organization in terms of pathways.
Given that I have used the swarm behavior of flocks of birds and Kauffman’s idea of cellular attractor’s as paradigmatic examples, one might wonder whether spontaneous self-organization is a necessary element of emergent complexity. It seems that this would exclude cases in which there is some kind of “systemic regulation”, i.e. cases where the system as a whole imposes constraints on the parts that shape its overall behavior.Footnote 11 Such cases of systemic control, however, are not excluded by my account of emergent complexity which invokes only the relationship between the effective complexity of the mechanism and the effective complexity of the behavior without requiring any specific organizational features. Thus, the issue is not so much about the presence or absence of control, but rather about how fine-grained the control is, if it is present. Yet, to say anything more determinate on this issue would likely require further clarification of the relevant notions of “control” and “self-organization” in this context.Footnote 12
Mechanical and emergent complexity seem to be two very different ideas of complexity in biology, but I have argued that they should be understood as two extremes of a spectrum. In particular, any real system is most likely situated somewhere between these two extremes. “Perfect” mechanical complexity would entail that the mechanism under study is actually the only member of its relevant ensemble. In order to provide a satisfactory explanation, one would have to describe it down to the smallest level of detail. But it is clear that even the most “mechanical” explanation in molecular biology must involve a lot of coarse-graining and treat many details of the underlying processes as irrelevant. At the other end of the spectrum, there does not seem to be any interesting emergent complexity if there is pure disorder and no regularities at all at the level of the mechanism.
My aim is not to take sides and argue that one of these versions of complexity is more relevant in biology than the other. Instead, I simply want to suggest that it is productive to think about biological complexity in terms of the more unifying framework I have proposed. This would also help us to overcome certain simplistic dichotomies, such as the idea that organisms can either be conceptualized as machines or as something completely different (cf. Nicholson 2013). My framework may also provide a helpful way of thinking about the relationship between mechanistic and so-called topological explanations, which has received considerable attention recently (e.g. Huneman 2010; Silberstein and Chemero 2013; Kostić 2018). The basic idea is that topological explanations explain by referring to structural properties at the level of the system (typically represented as a network) and “abstract away from the details of particular causal interactions” (Kostić 2018, p. 3). Similarly, Silberstein and Chemero argue that many explanations in neuroscience are based on “topological features (network architecture) of the network that are partially insensitive to and decoupled from and have a one-to-many relationship with respect to lower-level neurochemical and wiring details” (Silberstein and Chemero 2013, p. 963). But in my view this can be understood simply as a different way of saying that the relevant ensemble corresponding to the mechanism in question is large and contains many systems that greatly vary in their structural details. The larger the ensemble, the more it will seem that the relevant regularities are “abstract” and in some sense “non-mechanistic”. At the same time, as outlined before, in such scenarios there will often be an important role for mathematical tools in order to link the description of the system to the explanandum behavior. But this shouldn’t lead us to believe that we are suddenly dealing with an entirely different kind of explanation. In fact, as Huneman himself concedes: “one could also say that there is a continuum between two poles, one that consists in unraveling mechanisms without regard for any topological properties and one that is purely topological” (Huneman 2010, p. 225). The meaning of “topological properties” in this debate is often not very clear and varies between authors, but it seems plausible that in a broad sense they might be understood as precisely those properties that are used to describe the “large” ensembles figuring in explanations of systems that exhibit emergent complexity.
My framework also suggests an explication of the concept of “emergence” in biological explanations: emergence is present if the effective complexity of the behavior is high in comparison with the effective complexity of the underlying mechanism. It thus provides a clear way of interpreting the idea that “the system is more than the sum of its parts”. I do not pretend, of course, to have thereby addressed all the vexing issues that the concept of emergence raises in philosophy more generally.
As mentioned in the beginning of this article, philosophers and biologists often invoke complexity in arguments for pluralism (Mitchell 2003; Kellert et al. 2006). Based on my framework, I suggest that the soundness of such arguments will strongly depend on the version of complexity at play. Emergent complexity is related to (relatively) low complexity of mechanism and to descriptions that are so general that they may apply to very different kinds of systems. In fact this kind of “universality” is precisely one of the appeals of the idea of complexity science:
A vision shared by most researchers in complex systems is that certain intrinsic, perhaps even universal, features capture fundamental aspects of complexity in a manner that transcends specific domains” (Carlson and Doyle 2002, p. 2538).
This suggests that mechanical and emergent complexity actually pull in different directions as far as the question of pluralism is concerned. Philosophers invoking complexity must therefore be very clear about the kind of complexity they are referring to in their arguments.
In this article, I have mainly focused on molecular and cell biology at the expense of other important areas in which complexity plays a key role, such as developmental biology, or ecology. However, I hope that this discussion may illuminate or contribute to discussions around complexity in these other domains as well.
Notes
Note that there are other ways in which the term “epistemic complexity” has been used in the philosophical literature. Some people use it to refer to the external description of a system as given by the knowing subject (Bailly and Longo 2003), while others mean by it the knowledge embedded in a system (Kováč 2007; Dasgupta 2013). Rheinberger himself uses the term “experimental complexity” to refer to the complexity of the experimental landscape that emerges in parallel with scientists’ attempts to cope with the ontic complexity of the research object.
This is for now a purely terminological choice and does not presuppose any particular philosophical position on the concept of emergence.
Apart from emphasizing the relation to engineered mechanical systems, I use “mechanical” instead of “mechanistic” to distinguish this concept from the complexity of mechanism as introduced in Sect. 3.3.
Here, I have a maximally broad conception of “mechanism” in mind that encompasses basically any kind of causal structure underlying a phenomenon. In particular, I do not want to endorse any stronger requirements that are sometimes associated with accounts of mechanistic explanation, such as decomposability or functional localization (cf. Silberstein and Chemero 2013) because I do not want my account of complexity to be restricted to biological systems that are “well-behaved” in some sense.
For a formal treatment, the reader should consult Gell-Mann (2002).
I am not hereby suggesting that there is consensus on these norms in the philosophical literature.
The logistic map is an equation that recursively defines a sequence of numbers according to the formula \(x_{n+1} = rx_n(1-x_n)\). It can be interpreted as a very simple model of a growth process, but for certain choices of the parameter r it exhibits complex periodic or chaotic behavior. The Lorenz system is a system of three simple differential equations that has a set of chaotic solutions, the so-called Lorenz attractor.
One might think that \(c = c_B/c_M\) would be a more economical way of capturing the relationship. But then one would obtain the counterintuitive result that all mechanically complex systems have \(c\approx 1\). An additional, more technical, argument is that C should have the same unit (information content measured e.g. in bits) as CM and CB.
It has recently been argued that the notion of “pathway” should not simply be considered as a subspecies of “mechanism”, and that the two notions play different explanatory roles (Ross 2021). Given my broad understanding of “mechanism”, I will glance over these differences in the present context.
For a thorough criticism of Kauffman’s view on the evolution of gene regulatory networks, see Sansom (2011).
I thank an anonymous reviewer for raising this point.
On the notion of “control” in biology, see e.g. Stegmann (2014).
References
Adami, C. (2002). What is complexity? BioEssays, 24(12), 1085–1094. https://doi.org/10.1002/bies.10192.
Alberts, B., Johnson, A., Lewis, J., Morgan, D., Raff, M., Roberts, K., & Walter, P. (2015). Molecular biology of the cell (5th ed.). Garland Science.
Alon, U. (2020). An introduction to systems biology: Design principles of biological circuits (2nd ed.). CRC Press.
Badii, R., & Politi, A. (1999). Complexity: Hierarchical structures and scaling in physics. Cambridge University Press.
Bailly, F., & Longo, G. (2003). Objective and epistemic complexity in biology. In N. Singh (Ed.), Proceedings of the international conference on theoretical neurobiology (pp. 62–79). National Brain Research Centre.
Barabási, A. L., & Oltvai, Z. N. (2004). Network biology: Understanding the cell’s functional organization. Nature Reviews Genetics, 5(2), 101–113. https://doi.org/10.1038/nrg1272.
Bechtel, W. (2011). Mechanism and biological explanation. Philosophy of Science, 78(4), 533–557. https://doi.org/10.1086/661513.
Bechtel, W., & Richardson, R. C. (2010). Discovering complexity: Decomposition and localization as strategies in scientific research (2nd ed.). MIT Press. https://doi.org/10.7551/mitpress/8328.001.0001.
Bhalla, U. S., & Iyengar, R. (1999). Emergent properties of networks of biological signaling pathways. Science, 283(5400), 381–387. https://doi.org/10.1126/science.283.5400.381.
Bonner, J. T. (1988). The evolution of complexity by means of natural selection. Princeton University Press.
Carlson, J. M., & Doyle, J. (2002). Complexity and robustness. Proceedings of the National academy of Sciences of the United States of America, 99(suppl. 1), 2538–2545. https://doi.org/10.1073/pnas.012582499.
Craver, C. F. (2007). Explaining the brain. Oxford University Press.
Dasgupta, S. (2013). Epistemic complexity and the sciences of the artificial. In H. Andersen, D. Dieks, W. J. Gonzalez, T. Uebel, & G. Wheeler (Eds.), New challenges to philosophy of science (pp. 313–323). Springer. https://doi.org/10.1007/978-94-007-5845-2.
Gell-Mann, M. (2002). What is complexity? In A. Q. Curzio & M. Fortis (Eds.), Complexity and industrial clusters, contributions to economics (pp. 13–24). Physica-Verlag.
Gell-Mann, M., & Lloyd, S. (1996). Information measures, effective complexity, and total information. Complexity, 2, 44–52.
Gould, S. J. (1996). Full house. Harmony Books.
Green, S., Şerban, M., Scholl, R., Jones, N., Brigandt, I., & Bechtel, W. (2018). Network analyses in systems biology: New strategies for dealing with biological complexity. Synthese, 195(4), 1751–1777. https://doi.org/10.1007/s11229-016-1307-6.
Hooker, C. (2011). Introduction to philosophy of complex systems. Part A: Towards a framework for complex systems. In C. Hooker (Ed.), Philosophy of complex systems (Vol. 10, pp. 3–90). North-Holland. https://doi.org/10.1016/B978-0-444-52076-0.50001-8.
Huang, S., Eichler, G., Bar-Yam, Y., & Ingber, D. E. (2005). Cell fates as high-dimensional attractor states of a complex gene regulatory network. Physical Review Letters, 94(12), 128701. https://doi.org/10.1103/PhysRevLett.94.128701.
Huang, S., Ernberg, I., & Kauffman, S. (2009). Cancer attractors: A systems view of tumors from a gene network dynamics and developmental perspective. Seminars in Cell & Developmental Biology, 20(7), 869–876. https://doi.org/10.1016/j.semcdb.2009.07.003.
Huneman, P. (2010). Topological explanations and robustness in biological sciences. Synthese, 177(2), 213–245. https://doi.org/10.1007/s11229-010-9842-z.
Jørgensen, C., & Linding, R. (2010). Simplistic pathways or complex networks? Current Opinion in Genetics & Development, 20(1), 15–22. https://doi.org/10.1016/j.gde.2009.12.003.
Kaiser, M. I., Kronfeldner, M., & Meunier, R. (2014). Interdisciplinarity in philosophy of science. Journal for General Philosophy of Science, 45(1), 59–70. https://doi.org/10.1007/s10838-014-9269-1.
Kauffman, S. (2004). A proposal for using the ensemble approach to understand genetic regulatory networks. Journal of Theoretical Biology, 230(4), 581–590. https://doi.org/10.1016/j.jtbi.2003.12.017.
Kauffman, S. A. (1969). Metabolic stability and epigenesis in randomly constructed genetic nets. Journal of Theoretical Biology, 22(3), 437–467. https://doi.org/10.1016/0022-5193(69)90015-0.
Kauffman, S. A. (1976). Articulation of parts explanation in biology and the rational search for them. In M. Grene & E. Mendelsohn (Eds.), Topics in the philosophy of biology (pp. 245–263). Springer.
Kellert, S. H., Longino, H. E., & Waters, C. K. (2006). Introduction: The pluralist stance. In S. H. Kellert, H. E. Longino, & C. K. Waters (Eds.), Scientific pluralism (pp. vii–xxix). University of Minnesota Press.
Kolmogorov, A. N. (1963). On Tables of Random Numbers. Sankhyā: The Indian Journal of Statistics , Series A (1961-2002),25(4), 369–376.
Kostić, D. (2018). Mechanistic and topological explanations: An introduction. Synthese, 195(1), 1–10. https://doi.org/10.1007/s11229-016-1257-z.
Kováč, L. (2007). Information and knowledge in biology. Plant Signaling & Behavior, 2(2), 65–73.
Ladyman, J., Lambert, J., & Wiesner, K. (2013). What is a complex system? European Journal for Philosophy of Science, 3(1), 33–67. https://doi.org/10.1007/s13194-012-0056-8.
Ladyman, J., & Wiesner, K. (2020). What is a complex system? Yale University Press.
Lewis, J. (2003). Autoinhibition with transcriptional delay: A simple mechanism for the zebrafish somitogenesis oscillator. Current Biology, 13(16), 1398–1408. https://doi.org/10.1016/S0960-9822(03)00534-7.
Lloyd, S. (2001). Measures of complexity: A nonexhaustive list. IEEE Control Systems Magazine, 21(4), 7–8. https://doi.org/10.1109/MCS.2001.939938.
Lloyd, S., & Pagels, H. (1988). Complexity as thermodynamic depth. Annals of Physics, 188(1), 186–213. https://doi.org/10.1016/0003-4916(88)90094-2.
Machamer, P., Darden, L., & Craver, C. F. (2000). Thinking about mechanisms. Philosophy of Science, 67(1), 1–25. https://doi.org/10.1086/392759.
McAllister, J. W. (2003). Effective complexity as a measure of information content. Philosophy of Science, 70(2), 302–307. https://doi.org/10.1086/375469.
McShea, D. W. (2005). The evolution of complexity without natural selection, a possible large-scale trend of the fourth kind. Paleobiology, 31(sp5), 146–156. https://doi.org/10.1666/0094-8373(2005)031[0146:TEOCWN]2.0.CO;2.
Mitchell, S. D. (2003). Biological complexity and integrative pluralism. Cambridge University Press.
Nicholson, D. J. (2013). Organisms\(\ne \)Machines. Studies in History and Philosophy of Science Part C: Studies in History and Philosophy of Biological and Biomedical Sciences, 44(4), 669–678. https://doi.org/10.1016/j.shpsc.2013.05.014.
Parker, G. A., & Smith, J. M. (1990). Optimality theory in evolutionary biology. Nature, 348(6296), 27–33. https://doi.org/10.1038/348027a0.
Parrott, L. (2010). Measuring ecological complexity. Ecological Indicators, 10(6), 1069–1076. https://doi.org/10.1016/j.ecolind.2010.03.014.
Rescher, N. (1998). Complexity: A philosophical overview. Routledge.
Rheinberger, H. J. (1997). Experimental complexity in biology: Some epistemological and historical remarks. Philosophy of Science, 64, S245–S254. https://doi.org/10.1086/392604.
Ross, L. N. (2021). Causal concepts in biology: How pathways differ from mechanisms and why it matters. The British Journal for the Philosophy of Science, 72(1), 131–158. https://doi.org/10.1093/bjps/axy078.
Sansom, R. (2011). Ingenious genes. MIT Press.
Silberstein, M., & Chemero, A. (2013). Constraints on localization and decomposition as explanatory strategies in the biological sciences. Philosophy of Science, 80(5), 958–970. https://doi.org/10.1086/674533.
Simon, H. A. (1962). The architecture of complexity. Proceedings of the American Philosophical Society, 106(6), 467–482.
Singh, R. S., & Gupta, B. P. (2020). Genes and genomes and unnecessary complexity in precision medicine. npj Genomic Medicine, 5(1), 1–9. https://doi.org/10.1038/s41525-020-0128-1.
Smith, J. M., & Szathmary, E. (1995). The major transitions in evolution. Oxford University Press.
Sober, E. (2015). Ockha’ razors: A user’s manual. Cambridge University Press. https://doi.org/10.1017/CBO9781107705937.
Stegmann, U. (2014). Causal control and genetic causation. Noûs, 48(3), 450–465.
Strogatz, S. H. (2001). Exploring complex networks. Nature, 410(6825), 268–276. https://doi.org/10.1038/35065725.
Van Regenmortel, M. H. V. (2004). Reductionism and complexity in molecular biology. EMBO Reports, 5, 1016–1020.
von Dassow, G., Meir, E., Munro, E. M., & Odell, G. M. (2000). The segment polarity network is a robust developmental module. Nature, 406(6792), 188–92. https://doi.org/10.1038/35018085.
Weaver, W. (1948). Science and complexity. American Scientist, 36(4), 536–544.
Weinberg, R. A. (2014). The biology of cancer (2nd ed.). Garland Science.
Wimsatt, W. C. (1972). Complexity and organization. PSA: Proceedings of the Biennial Meeting of the Philosophy of Science Association, 1972, 67–86. https://doi.org/10.1086/psaprocbienmeetp.1972.3698961.
Wimsatt, W. C. (1974). Reductive explanation: A functional account. PSA: Proceedings of the Biennial Meeting of the Philosophy of Science Association, 1974, 671–710.
Wimsatt, W. C. (2007). Re-engineering philosophy for limited beings: Piecewise approximations to reality. Harvard University Press.
Acknowledgements
I would like to thank the members of the Conceptual Biology and Medicine Team at ImmunoConcEpT Bordeaux for useful feedback. In addition, I would like to thank Giovanni Boniolo for his encouragement.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article belongs to the topical collection “Simplicity out of Complexity? Physics and the Aims of Science”, edited by Florian J. Boge, Martin King, Paul Grünke and Miguel Ángel Carretero Sahuquillo.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gross, F. Between mechanical clocks and emergent flocks: complexities in biology. Synthese 199, 12073–12102 (2021). https://doi.org/10.1007/s11229-021-03324-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11229-021-03324-z