Abstract
Key message
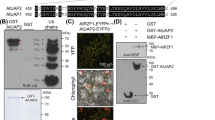
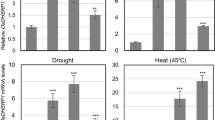
KEG is involved in mediating the proteasome-dependent degradation of FDH, a stress-responsive enzyme. The UPS may function to suppress FDH mediated stress responses under favorable growth conditions.
Abstract
Formate dehydrogenase (FDH) has been studied in bacteria and yeasts for the purpose of industrial application of NADH co-factor regeneration. In plants, FDH is regarded as a universal stress protein involved in responses to various abiotic and biotic stresses. Here we show that FDH abundance is regulated by the ubiquitin proteasome system (UPS). FDH is ubiquitinated in planta and degraded by the 26S proteasome. Interaction assays identified FDH as a potential substrate for the RING-type ubiquitin ligase Keep on Going (KEG). KEG is capable of attaching ubiquitin to FDH in in vitro assays and the turnover of FDH was increased when co-expressed with a functional KEG in planta, suggesting that KEG contributes to FDH degradation. Consistent with a role in regulating FDH abundance, transgenic plants overexpressing KEG were more sensitive to the inhibitory effects of formate. In addition, FDH is a phosphoprotein and dephosphorylation was found to increase the stability of FDH in degradation assays. Based on results from this and previous studies, we propose a model where KEG mediates the ubiquitination and subsequent degradation of phosphorylated FDH and, in response to unfavourable growth conditions, reduction in FDH phosphorylation levels may prohibit turnover allowing the stabilized FDH to facilitate stress responses.






Similar content being viewed by others
References
Abràmoff MD, Magalhães PJ, Ram SJ (2004) Image processing with imageJ. Biophotonics Int 11:36–41. https://doi.org/10.1117/1.3589100
Alekseeva A, Savin S, Tishkov V (2011) NAD (+) -dependent formate dehydrogenase from plants. Acta Nat 3:38–54
Bykova NV, Stensballe A, Egsgaard H, Jensen ON, Møller IM (2003) Phosphorylation of formate dehydrogenase in potato tuber mitochondria. J Biol Chem 278:26021–26030. https://doi.org/10.1074/jbc.M300245200
Chau V, Tobias JW, Bachmair A, Marriott D, David J, Gonda DK, Varshavsky A, Chau V, Tobias JW, Bachmair A, Marriott D, Eckert DJ, Gonda DK, Varshavsky A (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. 243:1576–1583
Choi DS, Kim NH, Hwang BK (2014) Pepper mitochondrial FORMATE DEHYDROGENASE1 regulates cell death and defense responses against bacterial pathogens. Plant Physiol 166:1298–1311. https://doi.org/10.1104/pp.114.246736
Colas des Francs-Small C, Ambard-Bretteville F, Small ID, Rémy R (1993) Identification of a major soluble protein in mitochondria from nonphotosynthetic tissues as NAD-dependent formate dehydrogenase. Plant Physiol 102:1171–1177. https://doi.org/10.1104/pp.102.4.1171
Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45:616–629. https://doi.org/10.1111/j.1365-313X.2005.02617.x
Finley RL, Zhang H, Zhong J, Stanyon CA (2002) Regulated expression of proteins in yeast using the MAL61-62 promoter and a mating scheme to increase dynamic range. Gene 285:49–57
Gu Y, Innes RW (2011) The keep on going protein of Arabidopsis recruits the enhanced disease resistance1 protein to trans-Golgi network/early endosome vesicles. Plant Physiol 155:1827–1838. https://doi.org/10.1104/pp.110.171785
Gu Y, Innes RW (2012) The KEEP ON GOING protein of Arabidopsis regulates intracellular protein trafficking and is degraded during fungal infection. Plant Cell 24:4717–4730. https://doi.org/10.1105/tpc.112.105254
Hanson AD, Roje S (2001) One-carbon metabolism in higher plants. Annu Rev Plant Physiol 52:119–137
Herman P, Ramberg H, Baack R (2002) Formate dehydrogenase in Arabidopsis thaliana: overexpression and subcellular localization in leaves. Plant Sci 163:1137–1145
Hourton-Cabassa C, Ambard-Bretteville F, Moreau F, Davy de Virville J, Rémy R, Francs-Small C (1998) Stress induction of mitochondrial formate dehydrogenase in potato leaves. Plant Physiol 116:627–635
Hunter T (2007) The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell 28:730–738. https://doi.org/10.1016/j.molcel.2007.11.019
Igamberdiev A, Bykova NV, Kleczkowski L (1999) Origins and metabolism of formate in higher plants. Plant Physiol Biochem 37:503–513
Kim D-Y, Scalf M, Smith LM, Vierstra RD (2013) Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 25:1523–1540. https://doi.org/10.1105/tpc.112.108613
Komander D, Rape M (2012) The ubiquitin code. Annu Rev Biochem 81:203–229. https://doi.org/10.1146/annurev-biochem-060310-170328
Kong L, Cheng J, Zhu Y, Ding Y, Meng J, Chen Z, Xie Q, Guo Y, Li J, Yang S, Gong Z (2015) Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat Commun 6:8630. https://doi.org/10.1038/ncomms9630
Kraft E, Stone S, Ma L, Su N, Gao Y, Lau O-S, Deng X-W, Callis J (2005) Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol 139:1597–1611. https://doi.org/10.1104/pp.105.067983.the
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Li R, Bonham-Smith PC, King J (2001) Molecular characterization and regulation of formate dehydrogenase in Arabidopsis thaliana. Can J Bot Can Bot 79:796–804. https://doi.org/10.1139/cjb-79-7-796
Li R, Moore M, Bonham-Smith P, King J (2002) Overexpression of formate dehydrogenase in Arabidopsis thaliana resulted in plants tolerant to high concentrations of formate. J Plant Physiol 159(10):1069–1076
Li R, Moore M, King J (2003) Investigating the regulation of one-carbon metabolism in Arabidopsis thaliana. Plant Cell Physiol 44:233–241
Liu H, Stone S (2010) Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22:2630–2641. https://doi.org/10.1105/tpc.110.076075
Liu H, Stone S (2013) Cytoplasmic degradation of the Arabidopsis transcription factor abscisic acid insensitive 5 is mediated by the RING-type E3 ligase KEEP ON GOING. J Biol Chem 288:20267–20279. https://doi.org/10.1074/jbc.M113.465369
Lorick KL, Jensen JP, Fang S, Ong a M, Hatakeyama S, Weissman AM (1999) RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA 96:11364–11369
Lou HQ, Gong YL, Fan W, Zhu JM, Liu Y, Cao MJ, Wang M-H, Yang JL, Zheng SJ (2016) VuFDH functions as a formate dehydrogenase that confers aluminum and low pH (H+) tolerance
Lyzenga WJ, Liu H, Schofield A, Muise-Hennessey A, Stone S (2013) Arabidopsis CIPK26 interacts with KEG, components of the ABA signalling network and is degraded by the ubiquitin-proteasome system. J Exp Bot 64:2779–2791. https://doi.org/10.1093/jxb/ert123
Olson B, Skavdahl M, Ramberg H, Osterman J, Markwell J (2000) Formate dehydrogenase in Arabidopsis thaliana: characterization and possible targeting to the chloroplast. Plant Sci 159:205–212
Patra B, Pattanaik S, Yuan L (2013) Ubiquitin protein ligase 3 mediates the proteasomal degradation of GLABROUS 3 and ENHANCER of GLABROUS 3, regulators of trichome development and flavonoid biosynthesis in Arabidopsis. Plant J 74:435–447. https://doi.org/10.1111/tpj.12132
Pauwels L, Ritter A, Goossens J, Durand AN, Liu H, Gu Y, Geerinck J, Boter M, Vanden Bossche R, De Clercq R, Van Leene J, Gevaert K, De Jaeger G, Solano R, Stone S, Innes RW, Callis J, Goossens A (2015) The RING E3 ligase KEEP ON GOING modulates JASMONATE ZIM-DOMAIN12 stability. Plant Physiol 169:1405–1417. https://doi.org/10.1104/pp.15.00479
Popov V, Lamzin V (1994) NAD + dependent formate dehydrogenase. Biochem J 301:625–643
Roitinger E, Hofer M, Köcher T, Pichler P, Novatchkova M, Yang J, Schlögelhofer P, Mechtler K (2015) Quantitative phosphoproteomics of the ATM and ATR dependent DNA damage response in Arabidopsis thaliana. Mol Cell Proteomics. https://doi.org/10.1074/mcp.M114.040352
Sawers G (1994) The hydrogenases and formate dehydrogenases of Escherichia coli. Antonie van Leeuwenhoek 66:57–88. https://doi.org/10.1007/BF00871633
Schwechheimer C, Deng X-W(2002) Studying protein-protein interactions with the yeast two-hybrid system. In: Gilmartin PM, Bowler C (eds) Molecular plant biology: a practical approach. Oxford University Press, Oxford
Serebriiskii IG, Toby GG, Finley RL, Golemis EA (2001) Genomic analysis utilizing the yeast two-hybrid system. Methods Mol Biol 175:415–454. https://doi.org/10.1038/nprot.2006.286
Sparkes I, Runions J, Kearns A, Hawes C (2006) Rapid, transient expression of fluorescent fusion proteinstobacco plants and generation of stably transformed plants. Nat Protoc 1:2019–2025.
Stone S, Williams L, Farmer L, Vierstra R, Callis J (2006) KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18:3415–3428. https://doi.org/10.1105/tpc.106.046532
Suzuki K, Itai R, Nakanishi H, Nishizawa NK, Yoshimura E, Mori S (1998) Formate dehydrogenase, an enzyme of anaerobic metabolism, is induced by iron deficiency in barley roots. Plant Physiol 116:725–732
Teng Y, Rezvani K, De Biasi M (2015) UBXN2A regulates nicotinic receptor degradation by modulating the E3 ligase activity of CHIP. Biochem Pharmacol 97:518–530. https://doi.org/10.1016/j.bcp.2015.08.084
Thacker JS, Yeung DH, Staines WR, Mielke JG (2016) Total protein or high-abundance protein: which offers the best loading control for Western blotting? Anal Biochem 496:76–78. https://doi.org/10.1016/j.ab.2015.11.022
Vierstra RD (2009) The ubiquitin–26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10:385–397. https://doi.org/10.1038/nrm2688
Wang F, Zhu D, Huang X, Li S, Gong Y, Yao Q, Fu X, Fan L-M, Deng XW (2009) Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell 21:2378–2390. https://doi.org/10.1105/tpc.108.065433
Wawrzynska A, Christiansen KM, Lan Y, Rodibaugh NL, Innes RW (2008) Powdery mildew resistance conferred by loss of the ENHANCED DISEASE RESISTANCE1 protein kinase is suppressed by a missense mutation in KEEP ON GOING, a regulator of abscisic acid signaling. Plant Physiol 148:1510–1522. https://doi.org/10.1104/pp.108.127605
Xue L, Wang P, Wang L, Renzi E, Radivojac P, Tang H, Arnold R, Zhu J-K, Tao WA (2013) Quantitative measurement of phosphoproteome response to osmotic stress in Arabidopsis based on Library-Assisted eXtracted Ion Chromatogram (LAXIC). Mol Cell Proteomics 53:1–39. https://doi.org/10.1017/CBO9781107415324.004
Acknowledgements
The authors would like to thank Dr. Russ Finley (Wayne State University) and Dr. Sonia Gazzarrini (University of Toronto) for providing the yeast two-hybrid vectors and cDNA library, respectively. This project was supported in part by a Natural Science and Engineering Research Council of Canada (NSERC) Discovery grant to S.L.S.
Author information
Authors and Affiliations
Contributions
DM: data acquisition, analysis of data and wrote the first draft. AS: data acquisition and editing of final draft. SLS conceived and designed the project, data interpretation and revisions of draft initial and subsequent drafts.
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2017_691_MOESM1_ESM.pdf
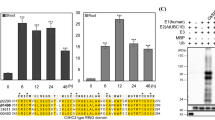
Supplemental Figure S1. Alignment of FDH amino acid sequences from 12 plant species showing predicted and experimentally determine sites of post-translational modification. Predicted ubiquitination (black outline) and phosphorylation (grey boxes) sites as well as experimentally determined phosphorylation (black arrowheads) sites are indicated. Plant species and associated accession/locus ID are as follows: Arabidopsis, Arabidopsis thaliana (At5g14780); Solanum, Solanum lycopersicum (tomato, NP_001234857); Oryza, Oryza sativa Japonica (rice, XP_015642621.1); Glycine,Glycine max (soybean, NP_001241141.1); Vitis,Vitis vinifera (grape, XP_002278444.1); Brachypodium, Brachypodium distachyon (stiff brome, XP_003563874); Sorghum, Sorghum bicolor (XP_002438408); Populus; Populus trichocarpa (XP_002320501.1); Selaginella, Selaginella moellendorffii (XP_002985142); Hordeum, Hordeum vulgare (barley, BAJ95739.1); Tuberosum, Solanum tuberosum (potato, NP_001274827.1); Capsicum,Capsicum annuum (sweet pepper, NP_001311723). Below the alignment asterisk (*) indicates fully conserved residues; colon (:) indicates amino acids that have highly similar properties; period (.) indicates amino acids that have weakly similar properties. Sequences were aligned using the Clustal Omega protein sequence alignment program available from The European Bioinformatics Institutes (EBI; http://www.ebi.ac.uk/) (McWilliam et al. 2013). (PDF 56 KB)
Rights and permissions
About this article
Cite this article
McNeilly, D., Schofield, A. & Stone, S.L. Degradation of the stress-responsive enzyme formate dehydrogenase by the RING-type E3 ligase Keep on Going and the ubiquitin 26S proteasome system. Plant Mol Biol 96, 265–278 (2018). https://doi.org/10.1007/s11103-017-0691-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-017-0691-8