Abstract
Key message
Expression differences underlie the functional differences between two related transcription factors: AINTEGUMENTA and AINTEGUMENTA-LIKE6. Ectopic expression of AINTEGUMENTA-LIKE6 at high levels alters floral organ initiation, growth and identity specification.
Abstract
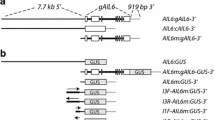
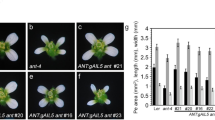
AINTEGUMENTA (ANT) and AINTEGUMENTA-LIKE6 (AIL6) encode related transcription factors with partially overlapping roles in floral organ development in Arabidopsis thaliana. To investigate whether the functional differences between ANT and AIL6 are a consequence of differences in gene expression and/or protein activity, we made transgenic plants in which a genomic copy of AIL6 was expressed under the control of the ANT promoter. ANT:gAIL6 can rescue the floral organ size defects of ant mutants when AIL6 is expressed at similar levels as ANT in wild type. Thus, the functional differences between ANT and AIL6 result primarily from gene expression differences. However, lines that express AIL6 at higher levels display additional phenotypes that include reduced numbers of floral organs and the production of mosaic floral organs. These phenotypes were also observed in two different inducible AIL6 transgenic lines but not in 35S:ANT, suggesting that AIL6 protein may have activities distinct from ANT, although the in vivo relevance of such differences is not clear. Similar to 35S:ANT plants, overexpression of AIL6 in the inducible lines also results in the production of larger flowers. The distinct phenotypes resulting from AIL6 misexpression in the transgenic lines described here and those previously characterized appear to result from different levels and patterns of AIL6 expression.









Similar content being viewed by others
References
Baker SC, Robinson-Beers K, Villanueva JM, Gaiser JC, Gasser CS (1997) Interactions among genes regulating ovule development in Arabidopsis thaliana. Genetics 145:1109–1124
Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad Sci Ser III Sci Vie 316:1194–1199
Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115:591–602
Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker, WQJ, Gerentes D, Perez P, Smyth DR (1996) AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8:155–168
Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B (2007) PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449:1053–1057
Gallegos JE, Rose AB (2015) The enduring mystery of intron-mediated enhancement. Plant Sci 237:8–15
Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM (2005) Patterns of auxin transport and gene expression during primodium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol 15:1899–1911
Ito T, Ng K-H, Lim T-S, Yu H, Meyerowitz EM (2007) The homeotic protein AGAMOUS controls late stamen development by regulating a jasmonate biosynthetic gene in Arabidopsis. Plant Cell 19:3516–3529
Jack T, Brockman LL, Meyerowitz EM (1992) The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68:683–687
Klucher KM, Chow H, Reiser L, Fischer RL (1996) The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8:137–153
Krizek BA (1999) Ectopic expression of AINTEGUMENTA in Arabidopsis plants results in increased growth of floral organs. Dev Genet 25:224–236
Krizek BA (2003) AINTEGUMENTA utilizes a mode of DNA recognition distinct from that used by proteins containing a single AP2 domain. Nucleic Acids Res 31:1859–1868
Krizek BA (2009) AINTEGUMENTA and AINTEGUMENTA-LIKE6 act redundantly to regulate Arabidopsis floral growth and patterning. Plant Physiol 150:1916–1929
Krizek BA (2011) Aintegumenta and Aintegumenta-Like6 regulate auxin-mediated flower development in Arabidopsis. BMC Res Notes 4:176
Krizek BA (2015a) AINTEGUMENTA-LIKE genes have partly overlapping functions with AINTEGUMENTA but make distinct contributions to Arabidopsis thaliana flower development. J Exp Bot 66:4537–4549
Krizek BA (2015b) Intronic sequences are required for AINTEGUMENTA-LIKE6 expression in Arabidopsis flowers. BMC Res Notes 8:556
Krizek BA, Eaddy M (2012) AINTEGUMENTA-LIKE6 regulates cellular differentiation in flowers. Plant Mol Biol 78:199–209
Krizek BA, Fletcher JC (2005) Molecular mechanisms of flower development: an armchair guide. Nat Rev Genet 6:688–698
Long J, Barton MK (2000) Initiation of axillary and floral meristems in Arabidopsis. Dev Biol 218:341–353
Mizukami Y, Fischer RL (2000) Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci USA 97:942–947
Nole-Wilson S, Krizek BA (2006) AINTEGUMENTA contributes to organ polarity and regulates growth of lateral organs in combination with YABBY genes. Plant Physiol 141:977–987
Nole-Wilson S, Tranby T, Krizek BA (2005) AINTEGUMENTA-like (AIL) genes are expressed in young tissues and may specify meristematic or division-competent states. Plant Mol Biol 57:613–628
Ptashne M (1988) How eukaryotic transcriptional activators work. Nature 225:683–689
Reinhardt D, Mandel T, Kuhlemeier C (2000) Auxin regulates the intiation and radial position of plant lateral organs. Plant Cell 12:507–518
Roslan HA, Salter MG, Wood CD, White, MRH, Croft KP, Robson F, Coupland G, Doonan J, Laufs P, Tomsett AB, Caddick MX (2001) Characterization of the ethanol-inducible alc gene-expression system in Arabidopsis thaliana. Plant J 28:225–235
Schultz, EA, Haughn, GW (1993) Genetic analysis of the floral initiation process (FLIP) in Arabidopsis. Development 119:745–765
Verwoerd TC, Dekker, BMM, Hoekema A (1989) A small scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res 17:2362
Weigel D, Nilsson O (1995) A developmental switch sufficient for flower initiation in diverse plants. Nature 377:495–500
Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69:843–859
Yamaguchi N, Jeong CW, Nole-Wilson S, Krizek BA, Wagner D (2016) AINTEGUMENTA and AINTEGUMENTA-LIKE6/PLETHORA3 induce LEAFY expression in response to auxin to promote the onset of flower formation in Arabidopsis. Plant Physiol 170:283–293
Yamaguchi N, Wu M-F, Winter CM, Berns MC, Nole-Wilson S, Yamaguchi A, Coupland G, Krizek BA, Wagner D (2013) A molecular framework for auxin-mediated initiation of flower primordia. Dev Cell 24:271–282
Acknowledgments
We thank Syngenta for the ALC switch plasmids (pJH0022 and pACN), Soumitra Ghoshroy and the Electron Microscopy Center staff for advice on the use of the SEM, and Jeff Twiss for use of the Leica TCS SP8X confocal microscope. This work was supported by National Science Foundation (NSF) Grants IOS 0922367 and 1354452.
Author contributions
BK conceived the study. HH and BK designed experiments, performed experiments and analyzed data. BK wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Han, H., Krizek, B.A. AINTEGUMENTA-LIKE6 can functionally replace AINTEGUMENTA but alters Arabidopsis flower development when misexpressed at high levels. Plant Mol Biol 92, 597–612 (2016). https://doi.org/10.1007/s11103-016-0535-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-016-0535-y