Abstract
Purpose
An understanding of higher order structure (HOS) of monoclonal antibodies (mAbs) could be critical to predicting its function. Amongst the various factors that can potentially affect HOS of mAbs, chemical modifications that are routinely encountered during production and long-term storage are of significant interest.
Methods
To this end, two Pfizer mAbs were subjected to forced deamidation stress for a period of eight weeks. Samples were aliquoted at various time points and high resolution accurate mass liquid chromatography-mass spectrometry (LC-MS/MS) was performed using low-artifact trypsin digestion (LATD) peptide mapping to identify and quantify chemical modifications. 2D backbone amide and sidechain methyl NMR spectra were acquired to gauge the effect of HOS changes upon chemical modification. Differential scanning calorimetry was also performed to assess the effect of thermal stability of mAbs upon modification. Finally, functional studies via target-binding based ELISA were performed to connect HOS changes to any loss of potency.
Results
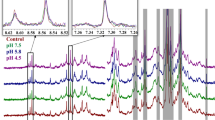
The extent of deamidation in the mAb domains were quantified by LC-MS/MS. The HOS changes as obtained from 2D NMR were mostly localized around the affected sites leaving the overall structure relatively unchanged. The antigen-antibody binding of the mAbs, in spite of deamidation in the Fab region, remains unchanged.
Conclusion
This case study provides an integrated approach of relating chemical modifications in mAb domains with possible changes in HOS. This can be potentially used to assess a possible loss of potency within the structure-function paradigm of proteins in an orthogonal manner.







Similar content being viewed by others
References
Kaplon H, Reichert JM. Antibodies to watch in 2018. mAbs. 2018;10(2):183–203.
Diepold K, Bomans K, Wiedmann M, Zimmermann B, Petzold A, Schlothauer T, et al. Simultaneous assessment of asp isomerization and Asn deamidation in recombinant antibodies by LC-MS following incubation at elevated temperatures. PLoS One. 2012;7(1):e30295.
Wang W, Singh S, Zeng DL, King K, Nema S. Antibody structure, instability, and formulation. J Pharm Sci. 2007;96:1–26.
Huang L, Lu J, Wroblewski VJ, Beals JM, Riggin RM. In vivo deamidation characterization of monoclonal antibody by LC/MS/MS. Anal Chem. 2005;77:1432–9.
Yan B, Steen S, Hambly D, Valliere-Douglass J, Bos TV, Smallwood S, et al. Succinimide formation at Asn 55 in the complementarity determining region of a recombinant monoclonal antibody IgG1 heavy chain. J Pharm Sci. 2009;98:3509–21.
Harris RJ, Kabakoff B, Macchi FD, Shen FJ, Kwong M, Andya JD, et al. Identification of multiple sources of charge heterogeneity in a recombinant antibody. J Chromatogr B Biomed Sci Appl. 2001;752:233–45.
Chu GC, Chelius D, Xiao G, Khor HK, Coulibaly S, Bondarenko PV. Accumulation of succinimide in a recombinant monoclonal antibody in mildly acidic buffers under elevated temperatures. Pharm Res. 2007;24:1145–56.
Pace AL, Wong RL, Zhang YT, Kao YH, Wang YJ. Asparagine deamidation dependence on buffer type, pH, and temperature. J Pharm Sci. 2013;102(6):1712–23.
Geiger T, Clarke S. Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. J Biol Chem. 1987;262(2):785–94.
Clarke S. Propensity for spontaneous succinimide formation from aspartyl and asparaginyl residues in cellular proteins. Int J Pept Protein Res. 1987;30(6):808–21.
Wright HT. Nonenzymatic deamidation of asparaginyl and glutaminyl residues in proteins. Crit Rev Biochem Mol Biol. 1991;26(1):1–52.
Robinson NE, Robinson AB. Molecular clocks. Proc Natl Acad Sci. 2001;98(3):944–9.
Kosky AA, Razzaq UO, Treuheit MJ, Brems DN. The effects of alpha-helix on the stability of Asn residues: deamidation rates in peptides of varying helicity. Protein Sci. 1999;8(11):2519–23.
Chelius D, Rehder DS, Bondarenko PV. Identification and characterization of deamidation sites in the conserved regions of human immunoglobulin gamma antibodies. Anal Chem. 2005;77(18):6004–11.
Robinson NE, Robinson AB. Deamidation of human proteins. Proc Natl Acad Sci. 2001;98(22):12409–13.
Oliyai C, Borchardt RT. Chemical pathways of peptide degradation. VI. Effect of the primary sequence on the pathways of degradation of aspartyl residues in model hexapeptides. Pharm Res. 1994;11(5):751–8.
Kossiakoff AA. Tertiary structure is a principal determinant to protein deamidation. Science. 1988;240(4849):191–4.
Athmer L, Kindrachuk J, Georges F, Napper S. The influence of protein structure on the products emerging from succinimide hydrolysis. J Biol Chem. 2002;277:30502–7.
Sydow JF, Lipsmeier F, Larraillet V, Hilger M, Mautz B, Mølhøj M, et al. Structure-based prediction of asparagine and aspartate degradation sites in antibody variable regions. PLoS One. 2014;9(6):e100736.
Vlasak J, Bussat MC, Wang S, Wagner-Rousset E, Schaefer M, Klinguer-Hamour C, et al. Identification and characterization of asparagine deamidation in the light chain CDR1 of a humanized IgG1 antibody. Anal Biochem. 2009;392:145–54.
Liu YD, van Enk JZ, Flynn GC. Human antibody fc deamidation in vivo. Biologicals. 2009;37(5):313–22.
Khawli LA, Goswami S, Hutchinson R, Kwong ZW, Yang J, Wang X, et al. Charge variants in IgG1: isolation, characterization, in vitro binding properties and pharmacokinetics in rats. MAbs. 2010;2:613–24.
Sinha S, Zhang L, Duan S, Williams TD, Vlasak J, Ionescu R, et al. Effect of protein structure on deamidation rate in the fc fragment of an IgG1 monoclonal antibody. Protein Sci. 2009;18:1573–84.
Geuijen KP, Oppers-Tiemissen C, Egging DF, Simons PJ, Boon L, Schasfoort R, et al. Rapid screening of IgG quality attributes–effects on fc receptor binding. FEBS open bio. 2017;7(10):1557–74.
Pande A, Mokhor N, Pande J. Deamidation of human γS-crystallin increases attractive protein interactions: implications for cataract. Biochemistry. 2015;54(31):4890–9.
Takata T, Oxford JT, Demeler B, Lampi KJ. Deamidation destabilizes and triggers aggregation of a lens protein, βA3-crystallin. Protein Sci. 2008;17(9):1565–75.
Luo, Q., Joubert, M. K., Stevenson, R., Ketchem, R. R., Narhi, L. O., & Wypych, J., Chemical modifications in therapeutic protein aggregates generated under different stress conditions. J Biol Chem, 2011. 286: p. 25134, 25144.
Bischoff R, Roecklin D, Roitsch C. Analysis of recombinant proteins by isoelectric focusing in immobilized pH gradients. Electrophoresis. 1992;13:214–9.
Groenen PJ, van Dongen MJ, Voorter CE, Bloemendal H, de Jong WW. Age-dependent deamidation of αB-crystallin. FEBS Lett. 1993;322:69–72.
Tsuji K, Baczynskyj L, Bronson GE. Capillary electrophoresis-electrospray mass spectrometry for the analysis of recombinant bovine and porcine somatotropins. Anal Chem. 1992;64:1864–70.
Aswad DW. Stoichiometric methylation of porcine adrenocorticotropin by protein carboxyl methyltransferase requires deamidation of asparagine 25. Evidence for methylation at the alpha-carboxyl group of atypical L-isoaspartyl residues. J Biol Chem. 1984;259:10714–21.
Teshima G, Stults JT, Ling V, Canova-Davis E. Isolation and characterization of a succinimide variant of methionyl human growth hormone. J Biol Chem. 1991;266:13544–7.
Lindner H, Sarg B, Grunicke H, Helliger W. Age-dependent deamidation of H1 histones in chromatin of mammalian tissues. J Cancer Res Clin Oncol. 1999;125:182–6.
Marzilli LA, Stephens E, Philip S, English AM, Rouse JC Minimizing method-induced deamidation and isomerization during antibody characterization to ensure optimal understanding of product quality attributes. LC-GC online. 2017;15(1):6–14.
Lehmann WD, Schlosser A, Erben G, Pipkorn R, Bossemeyer D, Kinzel V. Analysis of isoaspartate in peptides by electrospray tandem mass spectrometry. Protein Sci. 2000;9(11):2260–8.
Carvalho RN, Solstad T, Bjørgo E, Barroso JF, Flatmark T. Deamidations in recombinant human phenylalanine hydroxylase identification of labile asparagine residues and functional characterization of asn→ asp mutant forms. J Biol Chem. 2003;278:15142–52.
Jedrzejewski PT, Girod A, Tholey A, König N, Thullner S, Kinzel V, et al. A conserved deamidation site at asn 2 in the catalytic subunit of mammalian cAMP-dependent protein kinase detected by capillary LC-MS and tandem mass spectrometry. Protein Sci. 1998;7:457–69.
Johnson BA, Aswad DW. Optimal conditions for the use of protein L-isoaspartyl methyltransferase in assessing the isoaspartate content of peptides and proteins. Anal Biochem. 1991;192:384–91.
Houde D, Peng Y, Berkowitz SA, Engen JR. Post-translational modifications differentially affect IgG1 conformation and receptor binding. Mol Cell Proteomics. 2010;9:1716–28.
Zhang Z, Zhang A, Xiao G. Improved protein hydrogen/deuterium exchange mass spectrometry platform with fully automated data processing. Anal Chem. 2012;84:4942–9.
Zhang A, Hu P, MacGregor P, Xue Y, Fan H, Suchecki P, et al. Understanding the conformational impact of chemical modifications on monoclonal antibodies with diverse sequence variation using hydrogen/deuterium exchange mass spectrometry and structural modeling. Anal Chem. 2014;86:3468–75.
Jain T, Sun T, Durand S, Hall A, Houston NR, Nett JH, et al. Biophysical properties of the clinical-stage antibody landscape. Proc Natl Acad Sci. 2017;114(5):944–9.
Lu X, Nobrega RP, Lynaugh H, Jain T, Barlow K, Boland T, et al. Deamidation and isomerization liability analysis of 131 clinical-stage antibodies. MAbs. 2018:1–13.
Liu H, Gaza-Bulseco G, Chumsae C. Glutamine deamidation of a recombinant monoclonal antibody. Rapid Communications in Mass Spectrometry: An International Journal Devoted to the Rapid Dissemination of Up-to-the-Minute Research in Mass Spectrometry. 2008;22(24):4081–8.
Yan Y, Wei H, Fu Y, Jusuf S, Zeng M, Ludwig R, et al. Isomerization and oxidation in the complementarity-determining regions of a monoclonal antibody: a study of the modification–structure–function correlations by hydrogen–deuterium exchange mass spectrometry. Anal Chem. 2016;88(4):2041–50.
Brinson RG, Marino JP, Delaglio F, Arbogast LW, Evans RM, Kearsley A, et al. Enabling adoption of 2D-NMR for the higher order structure assessment of monoclonal antibody therapeutics. mAbs. 2018;11(1):94–105.
Hodgson DJ, Ghasriani H, Aubin Y. Assessment of the higher order structure of Humira®, Remicade®, Avastin®, Rituxan®, Herceptin®, and Enbrel® by 2D-NMR fingerprinting. J Pharm Biomed Anal. 2019;163:144–52.
Yagi H, Zhang Y, Yagi-Utsumi M, Yamaguchi T, Iida S, Yamaguchi Y, et al. Backbone 1 H, 13 C, and 15 N resonance assignments of the fc fragment of human immunoglobulin G glycoprotein. Biomolecular NMR assignments. 2015;9(2):257–60.
Lam XM, Yang JY, Cleland JL. Antioxidants for prevention of methionine oxidation in recombinant monoclonal antibody HER2. J Pharm Sci. 1997;86(11):1250–5.
Khor HK, Jacoby ME, Squier TC, Chu GC, Chelius D. Identification of methionine sulfoxide diastereomers in immunoglobulin gamma antibodies using methionine sulfoxide reductase enzymes. mAbs. 2010;2(3):299–308.
Sinha S, Zhang L, Duan S, Williams TD, Vlasak J, Ionescu R, et al. Effect of protein structure on deamidation rate in the Fc fragment of an IgG1 monoclonal antibody. Protein Sci. 2009;18(8):1573–84.
Latypov RF, Hogan S, Lau H, Gadgil H, Liu D. Elucidation of acid-induced unfolding and aggregation of human immunoglobulin IgG1 and IgG2 fc. J Biol Chem. 2011:jbc–M111.
Liu D, Ren D, Huang H, Dankberg J, Rosenfeld R, Cocco MJ, et al. Structure and stability changes of human IgG1 Fc as a consequence of methionine oxidation. Biochemistry. 2008;47(18):5088–100.
Arbogast LW, Brinson RG, Marino JP. Mapping monoclonal antibody structure by 2D 13C NMR at natural abundance. Anal Chem. 2015;87(7):3556–61.
Majumder S, Jones MT, Kimmel M, Ignatius AA. Probing conformational diversity of Fc domains in aggregation-prone monoclonal antibodies. Pharm Res. 2018;35(11):220.
Barb AW, Prestegard JH. NMR analysis demonstrates immunoglobulin G N-glycans are accessible and dynamic. Nat Chem Biol. 2011;7(3):147–53.
Yan Y, Wei H, Fu Y, Jusuf S, Zeng M, Ludwig R, et al. Isomerization and oxidation in the complementarity-determining regions of a monoclonal antibody: a study of the modification–structure–function correlations by hydrogen–deuterium exchange mass spectrometry. Anal Chem. 2016;88(4):2041–50.
Lawrence XY, Amidon G, Khan MA, Hoag SW, Polli J, Raju GK, et al. Understanding pharmaceutical quality by design. AAPS J. 2014;16(4):771–83.
Jia L, Sun Y. Protein asparagine deamidation prediction based on structures with machine learning methods. PLoS One. 2017;12(7):e0181347.
Ohkuri T, Murase E, Sun SL, Sugitani J, Ueda T. Characterization of deamidation at Asn138 in L-chain of recombinant humanized fab expressed from Pichia pastoris. The Journal of Biochemistry. 2013;154(4):333–40.
Amit AG, Mariuzza RA, Phillips SE, Poljak RJ. Three-dimensional structure of an antigen-antibody complex at 2.8 a resolution. Science. 1986;233(4765):747–53.
Peng, H.P., Lee, K. H., Jian, J. W., & Yang, A. S. , Origins of specificity and affinity in antibody–protein interactions. , 201401131. Proceedings of the National Academy of Sciences, 2014. 201401131.
Bonnington L, Lindner I, Gilles U, Kailich T, Reusch D, Bulau P. Application of hydrogen/deuterium exchange-mass spectrometry to biopharmaceutical development requirements: improved sensitivity to detection of conformational changes. Anal Chem. 2017;89(16):8233–7.
Huang L, Lu X, Gough PC, De Felippis MR. Identification of racemization sites using deuterium labeling and tandem mass spectrometry. Anal Chem. 2010;82(15):6363–9.
Falconer, D.J., & Barb, A. W. , Mouse IgG2c Fc loop residues promote greater receptor-binding affinity than mouse IgG2b or human IgG1. PLoS One, 2018. 13(2): p. e0192123.
Acknowledgments and Disclosures
SM and AAI acknowledge the support from the Pfizer Worldwide R&D postdoctoral program and the leadership team in Pfizer Biotherapeutics Pharmaceutical Sciences and Pharmaceutical R&D. The authors gratefully acknowledge Dr. Markus Voehler in scheduling and assisting with NMR experiments at the high resolution Biomolecular NMR facility at Vanderbilt University. Sincere thanks to Hai-Young Kim and David Cirelli for their critical review of the manuscript. The authors thank Jamie Lee and Jason Kennedy for generating ELISA data.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Figure S1
(PNG 133 kb)
Figure S2
(PNG 1248 kb)
Figure S3
(PNG 56 kb)
Figure S4
(PNG 254 kb)
Figure S5
(PNG 852 kb)
Figure S6
(PNG 337 kb)
Figure S7
(PNG 1033 kb)
Figure S8
(PNG 582 kb)
Figure S9
(PNG 54 kb)
Figure S10
(PNG 53 kb)
Figure S11
(PNG 517 kb)
Figure S12
(PNG 194 kb)
Figure S13
(PNG 547 kb)
Figure S14
(PNG 296 kb)
Figure S15
(PNG 271 kb)
ESM 1
(DOCX 19 kb)
ESM 1
(DOCX 19.3 kb)
Rights and permissions
About this article
Cite this article
Majumder, S., Saati, A., Philip, S. et al. Utility of High Resolution NMR Methods to Probe the Impact of Chemical Modifications on Higher Order Structure of Monoclonal Antibodies in Relation to Antigen Binding. Pharm Res 36, 130 (2019). https://doi.org/10.1007/s11095-019-2652-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11095-019-2652-1