Abstract
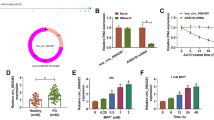
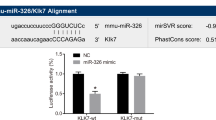
MicroRNA-93 (miR-93) is an oncogene that promotes tumor growth and angiogenesis. However, its role in Parkinson’s disease (PD) remains unknown. This study aimed at investigating the role of miR-93 in PD and the molecular mechanisms involved. 1-methyl-4-phenyl-1, 2, 3, 6-tetrahydropyridine (MPTP)-induced PD mouse model and lipopolysaccharide (LPS)-exposed BV2 cells were constructed. Real-time quantitative PCR was used to detect the mRNA expression of miR-93, iNOS, IL-6, IL-10, TNF-α and TGF-β1. Bioinformatics analysis and luciferase reporter assay were used to predict and confirm the interaction between miR-93 and STAT3. Flow cytometry was used to detect cell apoptosis. Western blotting was used to detect the protein expression of STAT3. Immunohistochemistry was used to analyze the Iba1-positive and TH positive cells. It was found that the expression of miR-93 was down-regulated in LPS-exposed BV2 cells. Overexpression of miR-93 inhibited the expression of iNOS, IL-6 and TNF-α, while enhanced the expression of TGF-β1 and IL-10. The expression of transcriptional activator 3 (STAT3) was found to be up-regulated in LPS-exposed BV2 cells. Knockdown of STAT3 inhibited the expression of iNOS, IL-6 and TNF-α, while enhanced the expression of TGF-β1 and IL-10. Moreover, STAT3 was found to be a direct target of miR-93, and miR-93 overexpression inhibited the expression of STAT3. Furthermore, both miR-93 overexpression and STAT3 knockdown reduced LPS-induced BV2 cell apoptosis, whereas STAT3 overexpression eliminated the inhibitory effect of miR-93 on LPS-induced BV2 cell apoptosis. In addition, miR-93 overexpression inhibited MPTP-induced STAT3 expression, microglial activation and inflammatory reaction and reduced the loss of tyrosine hydroxylase in the substantia nigra of mice. In conclusion, we demonstrate that miR-93 may be involved in PD by regulating the expression of STAT3.





Similar content being viewed by others
Data Availability
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
Abbreviations
- PD:
-
Parkinson’s disease
- MiRs:
-
MicroRNAs
- LPS:
-
Lipopolysaccharide
- STAT3:
-
Signal transductors and transcriptional activator 3
References
Hill-Burns EM et al (2017) Parkinson’s disease and Parkinson’s disease medications have distinct signatures of the gut microbiome. Mov Disord 32(5):739–749
Picca A et al (2020) Mitochondrial signatures in circulating extracellular vesicles of older adults with Parkinson’s disease: results from the EXosomes in PArkiNson’s Disease (EXPAND) study. J Clin Med 9(2):504
Burbulla LF et al (2017) Dopamine oxidation mediates mitochondrial and lysosomal dysfunction in Parkinson’s disease. Science 357(6357):1255–1261
Lill CM (2016) Genetics of Parkinson’s disease. Mol Cell Probes 30(6):386–396
Hoss AG et al (2016) microRNA profiles in Parkinson’s disease prefrontal cortex. Front Aging Neurosci 8:36
Cao X-Y et al (2017) MicroRNA biomarkers of Parkinson’s disease in serum exosome-like microvesicles. Neurosci Lett 644:94–99
Ding H et al (2016) Identification of a panel of five serum miRNAs as a biomarker for Parkinson’s disease. Parkinsonism Relat Disord 22:68–73
Thome AD et al (2016) microRNA-155 regulates alpha-synuclein-induced inflammatory responses in models of Parkinson disease. J Neurosci 36(8):2383–2390
Taguchi Y, Wang H (2018) Exploring MicroRNA biomarkers for Parkinson’s disease from mRNA expression profiles. Cells 7(12):245
Li S et al (2016) MicroRNA-7 inhibits neuronal apoptosis in a cellular Parkinson’s disease model by targeting Bax and Sirt2. Am J Transl Res 8(2):993
Yao L et al (2019) MicroRNA-124 regulates the expression of p62/p38 and promotes autophagy in the inflammatory pathogenesis of Parkinson’s disease. FASEB J 33(7):8648–8665
Fang L et al (2011) MicroRNA miR-93 promotes tumor growth and angiogenesis by targeting integrin-β8. Oncogene 30(7):806
Yang T et al (2016) Elevated serum miR-93, miR-191, and miR-499 are noninvasive biomarkers for the presence and progression of traumatic brain injury. J Neurochem 137(1):122–129
Choi D-J, Kwon J-K, Joe E-H (2018) A Parkinson’s disease gene, DJ-1, regulates astrogliosis through STAT3. Neurosci Lett 685:144–149
Zhang J et al (2019) miR-let-7a suppresses α-Synuclein-induced microglia inflammation through targeting STAT3 in Parkinson’s disease. Biochem Biophys Res Commun 519(4):740–746
Wang B et al (2016) Neuroprotective effects of nitidine in Parkinson’s disease models through inhibiting microglia activation: role of the Jak2–Stat3 pathway. RSC Adv 6(75):71328–71337
Yan A et al (2019) Idebenone alleviates neuroinflammation and modulates microglial polarization in LPS-stimulated BV2 cells and MPTP-induced Parkinson’s disease mice. Front Cell Neurosci 12:529
Hantraye P et al (1996) Inhibition of neuronal nitric oxide synthase prevents MPTP–induced Parkinsonism in baboons. Nat Med 2(9):1017–1021
Friedman BA et al (2018) Diverse brain myeloid expression profiles reveal distinct microglial activation states and aspects of Alzheimer’s disease not evident in mouse models. Cell Rep 22(3):832–847
Burguillos M et al (2011) Apoptosis-inducing factor mediates dopaminergic cell death in response to LPS-induced inflammatory stimulus: evidence in Parkinson’s disease patients. Neurobiol Dis 41(1):177–188
Dutta G, Zhang P, Liu B (2008) The lipopolysaccharide Parkinson’s disease animal model: mechanistic studies and drug discovery. Fundam Clin Pharmacol 22(5):453–464
Qin L et al (2005) Interactive role of the toll-like receptor 4 and reactive oxygen species in LPS-induced microglia activation. Glia 52(1):78–84
Zhou Y et al (2016) MicroRNA-7 targets Nod-like receptor protein 3 inflammasome to modulate neuroinflammation in the pathogenesis of Parkinson’s disease. Mol Neurodegener 11(1):28
Yao L et al (2018) MicroRNA-124 regulates the expression of MEKK3 in the inflammatory pathogenesis of Parkinson’s disease. J Neuroinflammation 15(1):13
Yan X-T et al (2017) MicroRNA-93 alleviates neuropathic pain through targeting signal transducer and activator of transcription 3. Int Immunopharmacol 46:156–162
Foshay KM, Gallicano GI (2009) miR-17 family miRNAs are expressed during early mammalian development and regulate stem cell differentiation. Dev Biol 326(2):431–443
Xiong L, Yu K, Zhen S (2018) MiR-93 blocks STAT3 to alleviate hepatic injury after ischemia–reperfusion. Eur Rev Med Pharmacol Sci 22(16):5295–5304
Yu H, Pardoll D, Jove R (2009) STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer 9(11):798–809
Bromberg J, Wang TC (2009) Inflammation and cancer: IL-6 and STAT3 complete the link. Cancer Cell 15(2):79–80
Liu M, Bing G (2011) Lipopolysaccharide animal models for Parkinson’s disease. Parkinson’s Dis. https://doi.org/10.4061/2011/327089
Forno L et al (1993) Similarities and differences between MPTP-induced parkinsonsim and Parkinson’s disease. Neuropathologic considerations. Adv Neurol 60:600–608
Acknowledgements
None.
Funding
None.
Author information
Authors and Affiliations
Contributions
ZL analyzed and interpreted the data, XW drafted the manuscript. FW trained the interviewers for data collection. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical Approval
Ethical clearance was obtained through the Ethics Review Committee, Faculty of Medicine, Union Hospital and the informed consent was obtained from all participants. Data collected from participants were kept confidential and were accessible only to the researchers.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, X., Liu, Z. & Wang, F. MicroRNA-93 Blocks Signal Transducers and Activator of Transcription 3 to Reduce Neuronal Damage in Parkinson’s Disease. Neurochem Res 46, 1859–1868 (2021). https://doi.org/10.1007/s11064-021-03333-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11064-021-03333-x