Abstract
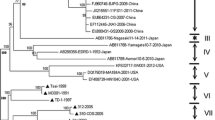
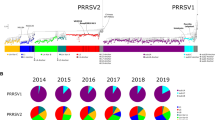
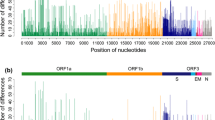
Porcine reproductive and respiratory syndrome virus (PRRSV) has been investigated extensively at the molecular level. Nevertheless, genome wide study on the temporal and spatial dynamics of the virus is non-existed. To explore this topic, we determined complete coding genome sequences for four PRRSV isolates and analyzed them together with 122 global published ones using the Bayesian coalescent approach as well as Bayesian inferences and maximum likelihood methods. All EU-type viruses belonged to one of two groups or were unclassified (5 isolates), and all NA-type isolates were divided into one of three major groups or were unclassified (1 isolate). Here, there was no apparent association between temporal or geographic origin and heterogeneity of global PRRSVs. Of the eight ORFs, ORF1a showed the most powerful evolutionary signal. Our findings also indicated that the PRRS virus evolved at a rate of 1.98 × 10−3 substitutions/site/year, and the most recent common ancestor of the virus existed 786.4 years ago. Here, EU-type viruses segregated 115.7 years ago, while NA-type isolates diverged 179.8 years before the present. In addition, our reconstruction of the effective population size depicted five phases of epidemic growth: an initial constant, followed sequentially by slow decrease, slight increase, sharp decline, and then a rapid expansion approaching the present.




Similar content being viewed by others
References
An TQ, Zhou YJ, Liu GQ, Tian ZJ, Li J, Qiu HJ, Tong GZ (2007) Genetic diversity and phylogenetic analysis of glycoprotein 5 of PRRSV isolates in mainland China from 1996 to 2006: coexistence of two NA-subgenotypes with great diversity. Vet Microbiol 123:43–52
Cha SH, Choi EJ, Park JH, Yoon SR, Song JY, Kwon JH, Song HJ, Yoon KJ (2006) Molecular characterization of recent Korean porcine reproductive and respiratory syndrome (PRRS) viruses and comparison to other Asian PRRS viruses. Vet Microbiol 117:248–257
Chen N, Cao Z, Yu X, Deng X, Zhao T, Wang L, Liu Q, Li X, Tian K (2011) Emergence of novel European genotype porcine reproductive and respiratory syndrome virus in mainland China. J Gen Virol 92:880–892
Darwich L, Gimeno M, Sibila M, Diaz I, de la Torre E, Dotti S, Kuzemtseva L, Martin M, Pujols J, Mateu E (2011) Genetic and immunological diversities of porcine reproductive and respiratory syndrome genotype I strains. Vet Microbiol 150:49–62
Dea S, Gagnon CA, Mardassi H, Pirzadeh B, Rogan D (2000) Current knowledge on the structural proteins of porcine reproductive and respiratory syndrome (PRRS) virus: comparison of the North American and European isolates. Arch Virol 145:659–688
Domingo E, Baranowski E, Escarmis C, Sobrino F (2002) Foot and mouth disease virus. Comp Immunol Microbiol Infect Dis 25:297–308
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214
Drummond AJ, Ho SY, Phillips MJ, Rambaut A (2006) Relaxed phylogenetics and dating with confidence. PLoS Biol 4:e88. doi:10.1371/journal.pbio.0040088
Duffy S, Shackelton LA, Holmes EC (2008) Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet 9:267–276
Elena SF, Sanjuan R (2005) Adaptive value of high mutation rates of RNA viruses: separating causes from consequences. J Virol 79:11555–11558
Forsberg R (2005) Divergence time of porcine reproductive and respiratory syndrome virus subtypes. Mol Biol Evol 22:2131–2134
Forsberg R, Oleksiewicz MB, Petersen AM, Hein J, Bøtner A, Storgaard T (2001) A molecular clock dates the common ancestor of European-type porcine reproductive and respiratory syndrome virus at more than 10 years before the emergence of disease. Virology 289:174–179
Guindon S, Gascuel O (2003) A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Hall TA (1999) BIOEDIT: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hanada K, Suzuki Y, Nakane T, Hirose O, Gojobori T (2005) The origin and evolution of porcine reproductive and respiratory syndrome viruses. Mol Biol Evol 22:1024–1031
Hao X, Lu Z, Kuang W, Sun P, Fu Y, Wu L, Zhao Q, Bao H, Fu Y, Cao Y, Li P, Bai X, Li D, Liu Z (2011) Polymorphic genetic characterization of the ORF7 gene of porcine reproductive and respiratory syndrome virus (PRRSV) in China. Virol J 8:73
Keffaber K (1989) Reproductive failure of unknown etiology. Am Assoc Swine Pract Newsl 1:1–10
Lauring AS, Andino R (2010) Quasispecies theory and the behavior of RNA viruses. PLoS Pathog 6:e1001005
Martin DP, Williamson C, Posada D (2005) RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21:260–262
Meulenberg JM (1998) Porcine reproductive and respiratory syndrome virus (PRRSV) molecular characterization of the agent. In: Proceedings of 15th IPVS congress, Birmingham, pp 149–157
Nam E, Park CK, Kim SH, Joo YS, Yeo SG, Lee C (2009) Complete genomic characterization of a European type 1 porcine reproductive and respiratory syndrome virus isolate in Korea. Arch Virol 154:629–638
Nelsen CJ, Murtaugh MP, Faaberg KS (1999) Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J Virol 73:270–280
Pesch S, Meyer C, Ohlinger VF (2005) New insights into the genetic diversity of European porcine reproductive and respiratory syndrome virus (PRRSV). Vet Microbiol 107:31–48
Plagemann PG (2003) Porcine reproductive and respiratory syndrome virus: origin hypothesis. Emerg Infect Dis 9:903–908
Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818
Rambaut A (2006) http://evolve.zoo.ox.ac.uk/beast
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Shi M, Lam TT, Hon CC, Hui RK, Faaberg KS, Wennblom T, Murtaugh MP, Stadejek T, Leung FC (2010) Molecular epidemiology of PRRSV: a phylogenetic perspective. Virus Res 154:7–17
Shi M, Lam TT, Hon C-C, Murtaugh MP, Davies PR, Hui RK, Li J, Wong LT, Yip CW, Jiang JW, Leung FC-C (2010) A phylogeny-based evolutionary, demographical and geographical dissection of North American type 2 porcine reproductive and respiratory syndrome viruses. J Virol 84:8700–8711
Snijder EJ, Meulenberg JJ (1998) The molecular biology of arteriviruses. J Gen Virol 79:961–979
Song J, Shen D, Cui J, Zhao B (2010) Accelerated evolution of PRRSV during recent outbreaks in China. Virus Genes 41:241–245
Stadejek T, Oleksiewicz MB, Potapchuk D, Podgorska K (2006) Porcine reproductive and respiratory syndrome virus strains of exceptional diversity in eastern Europe support the definition of new genetic subtypes. J Gen Virol 87:1835–1841
Suchard MA, Weiss RE, Sinsheimer JS (2001) Bayesian selection of continuous-time Markov chain evolutionary models. Mol Biol Evol 18:1001–1013
Suyama M, Torrents D, Bork P (2006) PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34:W609–W612
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal-windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 22:4673–4680
Wang C, Wu B, Amer S, Luo J, Zhang H, Guo Y, Dong G, Zhao B, He H (2010) Phylogenetic analysis and molecular characteristics of seven variant Chinese field isolates of PRRSV. BMC Microbiol 10:146
Wensvoort G, Terpstra C, Pol JM, ter Laak EA, Bloemraad M, de Kluyver EP, Kragten C, van Buiten L, den Besten A, Wagenaar F et al (1991) Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet Q 13:121–130
Yang Z (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13:555–556
Yoon SH, Song JY, Lee CH, Choi EJ, Cho IS, Kim B (2008) Genetic characterization of the Korean porcine reproductive and respiratory syndrome viruses based on the nucleocapsid protein gene (ORF7) sequences. Arch Virol 153:627–635
Zhu L, Zhang G, Ma J, He X, Xie Q, Bee Y, Gong SZ (2011) Complete genomic characterization of a Chinese isolate of porcine reproductive and respiratory syndrome virus. Vet Microbiol 147:274–282
Acknowledgments
This work was supported by a grant from the Next-Generation BioGreen 21 Program (Nos. PJ008196 and PJ009019), Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Sook Hee Yoon and Hyekwon Kim have contributed in equal measure to the work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yoon, S.H., Kim, H., Kim, J. et al. Complete genome sequences of porcine reproductive and respiratory syndrome viruses: perspectives on their temporal and spatial dynamics. Mol Biol Rep 40, 6843–6853 (2013). https://doi.org/10.1007/s11033-013-2802-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-013-2802-1