Abstract
Our data in the previous report demonstrated that C10orf116 (AFRO) is an adipocyte lineage-specific nuclear factor that can modulate the master adipogenesis transcription factors early during differentiation. However, more precise functional properties of this gene need to be clarified and await further investigation. Therefore, in this study, we performed an expression profile of cellular MicroRNAs (miRNAs) in the C10orf116 overexpression 3T3-L1 adipocytes and performed target prediction and functional enrichment of the differentially expressed miRNAs. Our study identified 34 miRNAs up-regulated in the 3T3-L1 adipocytes stably overexpressing C10orf116, whereas 43 miRNAs up-regulated in the control cells. The target genes of differentially expressed miRNAs were found to be involved in multiple signalling pathways, such as Wnt, TGF-beta, MAPK, Jak-STAT, insulin signalling pathway, et al. Our data provided novel information for the identification of C10orf116.
Similar content being viewed by others
Introduction
Obesity is a serious medical problem because it increases the risk of hypertension, type 2 diabetes mellitus, coronary heart disease, sleep-breathing disorders, certain forms of cancer [1], and it appears to markedly decrease life expectancy due to an increased risk of death by all causes [2]. The current epidemic of obesity in developed and developing nations, with its implications for public health, emphasizes the importance of understanding the complex mechanisms implicated in obesity development. Unfortunately, although over 600 genes, markers and chromosomal regions have been identified as associated with or linked to human obesity phenotypes [3], the responsible genes remain unknown in >95 % of severe obesity cases [4]. Thus, identification of novel genes and proteins associated with the development of obesity remains an important issue.
Our data in the previous report [5] demonstrated that C10orf116 is highly expressed in adipose tissue and is localized primarily within the nucleus. Overexpression studies in 3T3-L1 adipocytes indicated that it up-regulates the transcription levels of C/EBPα and PPARγ and promotes adipogenic differentiation starting from the early stage of adipogenesis. However, more precise functional properties of this gene need to be clarified and await further investigation.
MicroRNAs (miRNAs) are single-stranded and small noncoding RNA molecules of 18–24 nt in length that negatively regulate the expression of target genes in a post-transcriptional manner, by binding to messenger RNAs in a sequence-specific manner [6]. A single miRNA can regulate the expression of many target genes, and a target gene can also be regulated by several miRNAs [7]. These short molecules are conserved from worms to mammals; this high conservation of miRNAs sequences highlights the significance of their function. They contribute to the regulation of a variety of biological functions across diverse organisms such as apoptosis, proliferation, differentiation, development, cell cycle, stem cell maintenance, metabolism and hematopoiesis [7–10]. Recent studies reported that adipogenesis in humans and mice is also regulated by miRNAs, with several miRNAs being reported to have pro- or anti-adipogenic roles [11, 12] through repression of various genes, including transcriptional factors such as PPARγ, PPARα and KLF5 [13–16]. Therefore, miRNAs might also be implicated in the mechanisms of C10orf116 regulating adipogenesis.
Current methodologies have been developed and applied successfully in miRNA profiling, including miRNA microarrays [7]. The use of microarrays for global characterization of miRNA expression is becoming an increasingly important research tool [17, 18]. Therefore, we performed an expression profile of cellular miRNAs in the 3T3-L1 adipocytes stably overexpressing C10orf116 in this study. Then, we performed target prediction and functional enrichment of the differentially expressed miRNAs. Our data provided novel information for the identification of C10orf116.
Materials and methods
Cell culture and differentiation
3T3-L1 preadipocytes (ATCC, Manassas, VA) were maintained in Dulbecco’s medified Eagle’s medium with 10 % fetal calf serum (Biomedia, Boussens, France), 100 U/ml penicillin, and 50 μg/ml streptomycin (Life Technologies, Inc.) at 37 °C in 5 % CO2. 3T3-L1 cell lines that stably integrated either the pcDNA3.1Myc/HisB-C10orf116 plasmid or the empty vector were established as previously described [5]. To induce differentiation, 2-day post-confluent 3T3-L1 preadipocytes were exposed to the differentiation cocktail (100 μM methylisobutylxanthine, 0.25 μM dexamethasone, 1 μg/ml insulin). Two days later, cells were switched to medium containing 1 μg/ml insulin for 2 days. The cells were then switched back to DMEM containing only 10 % FCS until day 8. Cultures were replenished every 2 days.
Total RNA preparation
RNA was isolated using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
MicroRNA microarray assay
Expression levels of 611 microRNAs (Sanger miRBase 12.0) were studied using μParaflo™ microRNA microarray kit (LC Sciences, Houston, TX, USA). The assay started from a 2 to 5 μg total RNA sample, which was size fractionated using a YM-100 Microcon centrifugal filter (Millipore, Billerica, MA, USA) and the small RNAs (<300 nt) isolated were 3′-extended with a poly(A) tail using poly(A) polymerase. An oligonucleotide tag was then ligated to the poly(A) tail for later fluorescent dye staining. Two different tags were used for the two RNA samples in dual-sample experiments. Hybridization was carried out overnight on an μParaflo™ microfluidic chip (LC Sciences, Houston, TX, USA) using a microcirculation pump (Atactic Technologies Inc., Houston, TX, USA) [19, 20]. On the microfluidic chip, each detection probe consisted of a chemically modified nucleotide-coding segment complementary to target miRNA (from miRBase, Version 10.0, http://microrna.sanger.ac.uk/sequences/) and a spacer segment of polyethylene glycol to extend the coding segment away from the substrate. The detection probes were made by in situ synthesis using photogenerated reagent (PGR) chemistry. The hybridization melting temperatures were balanced by chemical modifications of the detection probes. Hybridization used 100 μL 6 × SSPE buffer (0.90 M NaCl, 60 mM Na2HPO4, 6 mM EDTA, pH 6.8) containing 25 % formamide at 34 °C. After hybridization detection by fluorescence labeling using tag-specific Cy3 (for cells transfected with the C10orf116-pcDNA3.1 vector) and Cy5 dyes (for cells transfected with the pcDNA.3.1 vector), hybridization images were collected using a laser scanner (GenePix 4000B; Molecular Devices, Union City, CA, USA) and digitized using Array-Pro image analysis software (Media Cybernetics, Bethesda, MD, USA).
Data were analyzed first by subtracting the background and then normalizing the signals using a locally-weighted regression (LOWESS) filter [21]. For two color experiments, the ratio of the two sets of detected signals (log2 transformed, balanced) and P values of the t test were calculated; differentially detected signals were those with less than 0.01 P values.
Quantitative real-time PCR
Quantitative real-time PCR (qRT-PCR) was performed with a miScript System (Qiagen, USA), which included specific primers for miRNAs. All procedures were performed according to the instructions provided by the manufacturer. The reverse transcription (RT) reaction system contained 1 μl miScript Reverse Transcriptase Mix, 4 μl 5× miScript RT Buffer and 15 μl RNase-free water. The 20 μl RT product was diluted into 100 μl. The procedures was 37 °C for 60 min and 95 °C for 5 min. Reaction system of quantitative Real-time PCR contained 10 μl SYBR Green PCR Master Mix, 2 μl miScript universal primer, 2 μl specific primer (mmu-miR-1944 CTC TGT GCT GAA TGT CAA GTT CUG ATT; U6 Sense: 5′-CGC TTC GGC AGC ACA TAT AC-3′ Antisense: 5′-TTC ACG AAT TTG CGT GTC AT-3′), 1 μl cDNA and 5 μl RNase-free water. The procedure for PCR was 95 °C 15 min; 94 °C 15 s, 55 °C 30 s, 70 °C 30 s, 45 cycles. RT reaction was performed in ABI 9700 PCR system (ABI, USA) and real-time PCR was performed in Rotor-gene 6000 PCR system (Corbett, Australia). Each reaction was run in duplicate and performed at least twice. Negative control reactions without RT reaction and template were also performed. Finally, the relative miR-1944 level was normalized to the endogenous control U6 expression was calculated by the 2−∆Ct method.
Bioinformatic analyses for mRNA target and molecular pathway prediction
Putative target genes regulated by the differentially expressed miRNAs were predicted bioinformatically and combining the prediction of their supposed targets with the bibliographic information of adipocyte gene expression. For this purpose TargetScan, MicroCosm and miRanda were used.
In order to identify molecular pathways potentially altered by the expression of multiple miRNAs, we used the DIANA-mirPath web-based computational tool [22]. The software performs an enrichment analysis of multiple miRNA target genes comparing each set of miRNA targets to all known KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways [23]. The combinatorial effect of co-expressed miRNAs in the modulation of a given pathway is taken into account by the simultaneous analysis of multiple miRNAs. The graphical output of the program provides an overview of the parts of the pathway modulated by microRNAs, facilitating the interpretation and presentation of the analysis results.
Results
MiRNA expression profiles in the 3T3-L1 adipocytes stably overexpressing C10orf116 relative to the 3T3-L1 adipocytes transfected with pcDNA 3.1
We used a miRNA microarray to evaluate miRNA expression profiles between the 3T3-L1 adipocytes stably overexpressing C10orf116 and transfected with pcDNA 3.1. The miRNA expression pattern was found to be different and only miRNAs with a statistically significant differential expression (P < 0.01) were listed (Fig. 1; Table 1). 34 miRNAs were found to be highly expressed in the 3T3-L1 adipocytes stably overexpressing C10orf116, whereas 43 miRNAs had lower expression compared to controls. Two of them, miR-712* and miR-1944, were significantly up-regulated with a fold change <−2, whereas miR-1907, miR-374, and miR-15b were significantly down-regulated with a fold change >2 in the 3T3-L1 adipocytes stably overexpressing C10orf116.
Validation of the microarrays results by qRT-PCR
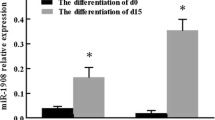
To further verify and validate the results obtained by microarrays, we performed qRT-PCR analysis of miR-1944. The result of qRT-PCR was shown in Fig. 2. The expression level of miR-1944 was significantly up-regulated in the 3T3-L1 adipocytes stably overexpressing C10orf116. The expression pattern obtained by qRT-PCR reflected the results obtained by microarrays, demonstrating the low false positive rate associated with microarrays in this experiment.
qRT-PCR analysis of miR-1944. qRT-PCR analysis was performed to quantify miR-1944 which was differentially expressed in the 3T3-L1 adipocytes stably overexpressing C10orf116 relative to the 3T3-L1 adipocytes transfected with pcDNA 3.1. Results were expressed as mean ± standard deviation (SD). * P < 0.05
Target prediction and functional analysis of differentially expressed miRNAs
To pursue the study, we performed a bioinformatic prediction in order to know the putative target genes regulated by all miRNAs differentially expressed in the 3T3-L1 adipocytes stably overexpressing C10orf116 and transfected with pcDNA 3.1. For this purpose, the following computational tools were used: TargetScan, MicroCosm and miRanda. TargetScan provides miRNA target predictions based on sequence complementary to target sites with emphasis on perfect base-pairing in the seed region and sequence conservation [24]. MicroCosm uses a weighted scoring system and rewards complementarity at the 5′ end of the microRNA [25]. miRanda computes optimal sequence complementarity between a set of mature microRNAs and a given mRNA using a weighted dynamic programming algorithm [9]. These web-based computational tools are based on different algorithms which are based on several parameters calculated individually for each miRNA. All the computational programs predicted potential target mRNA genes for the 5 miRNAs significantly differentially expressed in the 3T3-L1 adipocytes stably overexpressing C10orf116 and the controls. For miR-712*, TargetScan predicted 0, MicroCosm predicted 751 and miRanda predicted 1224 target genes. For miR-1944, TargetScan predicted 0, MicroCosm predicted 0 and miRanda predicted 8626 target genes. For miR-1907, TargetScan predicted 1020, MicroCosm predicted 0 and miRanda predicted 8081 target genes. For miR-374, TargetScan predicted 532, MicroCosm predicted 754 and miRanda predicted 6223 target genes. For miR-15b, TargetScan predicted 1020, MicroCosm predicted 977 and miRanda predicted 8076 target genes (Fig. 3).
In order to obtain a list of pathways likely to be specifically controlled by miRNAs significantly differentially expressed in the C10orf116 overexpression 3T3-L1 adipocytes and the controls, we performed an analysis of the most common signalling pathways by using the web-based computational tool named DIANA-miRPath. This computational tool estimates the impact of co-expressed miRNAs in biological pathways. Table 2 represents the DIANA-miRPath analysis based on TargetScan 5 target prediction software. As shown in Table 2, genes targeted by only miR-374 and miR-15b were found to be involved in lipid metabolism pathways, such as the Wnt signaling pathway, MAPK signaling pathway, Jak-STAT signaling pathway, etc.
Discussion
Obesity is an increasing global health problem that is usually accompanied by several significant health impairments, such as type 2 diabetes or cardiovascular disease [26]. Considerable evidence shows that obesity is caused by the interaction between multiple genes and the environment [27, 28]. A better understanding of the candidate genes required for the development of obesity could form the basis for new therapies that directly target molecular mechanisms underlying obesity. In the previous report [5], we identified C10orf116 as a novel gene that might play a role in obesity. Over-expression studies in 3T3-L1 cells indicated that it up-regulates the transcription levels of C/EBPα and PPARγ and promotes adipogenic differentiation starting from the early stage of adipogenesis. In the present study, to better understand the precise functional properties of the novel gene C10orf116 in obesity and to investigate a possible role of miRNAs in C10orf116-related genes regulation, we isolated and analyzed miRNAs in the 3T3-L1 adipocytes stably overexpressing C10orf116 relative to the 3T3-L1 adipocytes transfected with pcDNA 3.1 using miRNA microarray analysis.
Our study identified 34 miRNAs up-regulated in the 3T3-L1 adipocytes stably overexpressing C10orf116, whereas 43 miRNAs had lower expression compared to controls. Two of them, miR-712* and miR-1944, were significantly up-regulated with a fold change <−2, whereas miR-1907, miR-374, and miR-15b were significantly down-regulated with a fold change >2 in the 3T3-L1 adipocytes stably overexpressing C10orf116.
Our profiling results were further validated by the detection of miR-1944 by qRT-PCR. Our microarray analyses showed that the relative expression levels for miR-1944 were 2742.79 for the 3T3-L1 adipocytes stably overexpressing C10orf116 and 480.29 for the control cells. Moreover, qPCR experiments showed that this miRNA was also up-regulated in the C10orf116 overexpression 3T3-L1 adipocytes, in particular 1.67 fold. The result demonstrated the good correlation associated with microarrays in this experiment.
Although many computational approaches have been developed to predict miRNA targets using sequence information, their accuracy is limited [24, 29–31]. To increase reliability, we used three web-based target prediction databases, TargetScan, MicroCosm, and miRanda. The numbers of targets for the significantly differentially expressed 5 miRNAs predicted from three algorithms were showed in Fig. 3. The key molecular pathways potentially altered by the 5 miRNAs were predicted by the DIANA-mirPath web-based computational tool. However, as shown in Table 2, only the target genes of miR-374 and miR-15b were found.
We observed that several targets of miR-374 and miR-15b are involved in Wnt signaling pathway. These included Apc, Ppp2r2c, Wnt5b, Fzd5, Axin2, Wnt5a, Lrp6, which are targets of miR-374; Ccnd3, Wnt3a, Wnt7a, Ppp2r1b, Nkd1, Smad2, Axin2, Ccnd2, Ppp2r1a, Fosl1, Lrp6, Btrc, Ccnd1, Smad3, which are targets of miR-15b. Our data showed that miR-374 and miR-15b were significantly down-regulated in the 3T3-L1 adipocytes stably overexpressing C10orf116. Therefore, Wnt signaling pathway should be activated by C10orf116. Wnt signaling has been demonstrated to be a molecular switch that control adipogenesis by maintaining preadipocytes in an undifferentiated state through inhibition of C/EBPα and PPARγ expression. However, conversely, we identified C10orf116 overexpression in 3T3-L1 adipocytes up-regulates the transcription levels of C/EBPα and PPARγ and promotes adipogenic differentiation in the previous report [5]. Understanding the real relatioanship between C10orf116 and Wnt signaling pathway, by which C10orf116 regulates adipocyte differentiation remains an important area for future research.
Furthermore, the target genes of miR-374 and miR-15b were also found to be involved in multiple signal pathways, such as the TGF-beta, MAPK, Jak-STAT, insulin signalling pathway, et al. MiRNA profiles obtained in this study might help elucidate the regulatory mechanisms that C10orf116 involved in obesity. However, although some algorithms were used to predict potential mRNA targets, only a few miRNAs could be validated and assigned to specific mRNAs. Our analysis is limited because these two miRNAs were both down-regulated in the C10orf116 overexpression 3T3-L1 adipocytes. Targets of other modulated miRNAs up-regulated that have not be confirmed might also regulate biological processes of cells. Therefore, future investigations are needed.
References
Kopelman PG (2000) Obesity as a medical problem. Nature 404(6778):635–643. doi:10.1038/35007508
Calle EE, Thun MJ, Petrelli JM, Rodriguez C, Heath CW Jr (1999) Body-mass index and mortality in a prospective cohort of U.S. adults. N Eng J Med 341(15):1097–1105. doi:10.1056/NEJM199910073411501
Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, Perusse L, Bouchard C (2006) The human obesity gene map: the 2005 update. Obesity (Silver Spring) 14(4):529–644. doi:10.1038/oby.2006.71
Flier JS (2004) Obesity wars: molecular progress confronts an expanding epidemic. Cell 116(2):337–350
Ni Y, Ji C, Wang B, Qiu J, Wang J, Guo X (2012) A Novel pro-adipogenesis factor abundant in adipose tissues and over-expressed in obesity acts upstream of PPARgamma and C/EBPalpha. J Bioenerg Biomembr. doi:10.1007/s10863-012-9492-6
He L, Hannon GJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5(7):522–531. doi:10.1038/nrg1379
Li W, Ruan K (2009) MicroRNA detection by microarray. Anal Bioanal Chem 394(4):1117–1124. doi:10.1007/s00216-008-2570-2
Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EK (2008) Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthr Res Therapy 10(4):R101. doi:10.1186/ar2493
Betel D, Wilson M, Gabow A, Marks DS, Sander C (2008) The microRNA.org resource: targets and expression. Nucleic Acids Res 36 (Database issue):D149–153. doi:10.1093/nar/gkm995
Jones SW, Watkins G, Le Good N, Roberts S, Murphy CL, Brockbank SM, Needham MR, Read SJ, Newham P (2009) The identification of differentially expressed microRNA in osteoarthritic tissue that modulate the production of TNF-alpha and MMP13. Osteoarthr Cartil/OARS Osteoarthr Res Soc 17(4):464–472. doi:10.1016/j.joca.2008.09.012
Xie H, Sun L, Lodish HF (2009) Targeting microRNAs in obesity. Expert Opin Ther Targets 13(10):1227–1238. doi:10.1517/14728220903190707
Romao JM, Jin W, Dodson MV, Hausman GJ, Moore SS, Guan LL (2011) MicroRNA regulation in mammalian adipogenesis. Exp Biol Med (Maywood) 236(9):997–1004. doi:10.1258/ebm.2011.011101
Lin Q, Gao Z, Alarcon RM, Ye J, Yun Z (2009) A role of miR-27 in the regulation of adipogenesis. FEBS J 276(8):2348–2358
Kim SY, Kim AY, Lee HW, Son YH, Lee GY, Lee JW, Lee YS, Kim JB (2010) miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARgamma expression. Biochem Biophys Res Commun 392(3):323–328. doi:10.1016/j.bbrc.2010.01.012
Martinelli R, Nardelli C, Pilone V, Buonomo T, Liguori R, Castano I, Buono P, Masone S, Persico G, Forestieri P, Pastore L, Sacchetti L (2010) miR-519d overexpression is associated with human obesity. Obesity (Silver Spring) 18(11):2170–2176. doi:10.1038/oby.2009.474
Kinoshita M, Ono K, Horie T, Nagao K, Nishi H, Kuwabara Y, Takanabe-Mori R, Hasegawa K, Kita T, Kimura T (2010) Regulation of adipocyte differentiation by activation of serotonin (5-HT) receptors 5-HT2AR and 5-HT2CR and involvement of microRNA-448-mediated repression of KLF5. Mol Endocrinol 24(10):1978–1987. doi:10.1210/me.2010-0054
Sarkar D, Parkin R, Wyman S, Bendoraite A, Sather C, Delrow J, Godwin AK, Drescher C, Huber W, Gentleman R, Tewari M (2009) Quality assessment and data analysis for microRNA expression arrays. Nucleic Acids Res 37(2):e17. doi:10.1093/nar/gkn932
Liang Y, Ridzon D, Wong L, Chen C (2007) Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 8:166. doi:10.1186/1471-2164-8-166
Gao X, Gulari E, Zhou X (2004) In situ synthesis of oligonucleotide microarrays. Biopolymers 73(5):579–596. doi:10.1002/bip.20005
Zhu Q, Hong A, Sheng N, Zhang X, Matejko A, Jun KY, Srivannavit O, Gulari E, Gao X, Zhou X (2007) microParaflo biochip for nucleic acid and protein analysis. Methods Mol Biol 382:287–312
Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19(2):185–193
Papadopoulos GL, Alexiou P, Maragkakis M, Reczko M, Hatzigeorgiou AG (2009) DIANA-mirPath: integrating human and mouse microRNAs in pathways. Bioinformatics 25(15):1991–1993. doi:10.1093/bioinformatics/btp299
Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28(1):27–30
Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB (2003) Prediction of mammalian microRNA targets. Cell 115(7):787–798
Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36 (Database issue):D154–D158. doi:10.1093/nar/gkm952
Mokdad AH, Ford ES, Bowman BA, Dietz WH, Vinicor F, Bales VS (2001) Marks JS (2003) Prevalence of obesity, diabetes, and obesity-related health risk factors. JAMA J Am Med Assoc 289(1):76–79
Lee YS (2009) The role of genes in the current obesity epidemic. Ann Acad Med Singapore 38(1):43–45
Walley AJ, Asher JE, Froguel P (2009) The genetic contribution to non-syndromic human obesity. Nat Rev Genet 10(7):431–442. doi:10.1038/nrg2594
John B, Sander C, Marks DS (2006) Prediction of human microRNA targets. Methods Mol Biol 342:101–113. doi:10.1385/1-59745-123-1:101
Maziere P, Enright AJ (2007) Prediction of microRNA targets. Drug Discovery Today 12(11–12):452–458. doi:10.1016/j.drudis.2007.04.002
Wang X, El Naqa IM (2008) Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics 24(3):325–332. doi:10.1093/bioinformatics/btm595
Acknowledgments
This work was supported by Grants from the National Key Basic Research Program of China (2013CB530604), the National Natural Science Foundation of China (No 81000349), and the Science and Technology Development Fund of Nanjing Medical University (09njmum052).
Author information
Authors and Affiliations
Corresponding author
Additional information
Jie Qiu and Xiao-guang Zhou have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Qiu, J., Zhou, Xg., Zhou, Xy. et al. Characterization of microRNA expression profiles in 3T3-L1 adipocytes overexpressing C10orf116. Mol Biol Rep 40, 6469–6476 (2013). https://doi.org/10.1007/s11033-013-2762-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-013-2762-5