Abstract
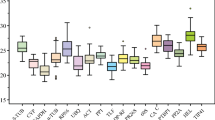
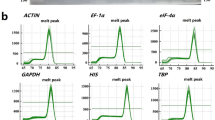
Quantitative real-time polymerase chain reaction (qRT-PCR) has been extensively used in several plant species as an accurate technique for gene expression analysis. However, the expression level of a target gene may be misconstrued due to unstable expression of the reference genes under different experimental conditions. Therefore, it is necessary to systematically evaluate these reference genes before experiments are conducted. Recently, more and more studies have focused on gene expression in pepper (Capsicum annuum L.). In this study, ten putative reference genes were chosen to identify expression stability by using geNorm and NormFinder statistical algorithms in ten different pepper sample pools, including those from different plant tissues (root, stem, leaf and flower) and from plants treated with hormones (salicylic acid and gibberellic acid) and abiotic stresses (cold, heat, salt and drought). EF1α and UEP exhibited the most stable expression across all of the tested pepper samples. For abiotic stress or different hormone treatment, the ranking of candidate reference genes was not completely consistent, except for EF1α which showed a relatively stable expression level. For different tissues, the expression of Actin1 was stable and it was considered an appropriate reference gene. It is concluded that EF1α, UEP and Actin1 are suitable reference genes for reliable qRT-PCR data normalization for the tissues and experimental conditions used in this experiment.


Similar content being viewed by others
References
Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250
Bustin SA, Benes V, Nolan T, Pfaffl MW (2005) Quantitative realtime RT-PCR—a perspective. J Mol Endocrinol 34:597–601
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17
Dilger M, Felsenstein FG, Schwarz G (2003) Identification and quantitative expression analysis of genes that are differentially expressed during conidial germination in Pyrenophora teres. Mol Gen Genomics 270:147–155
Doshi KM, Eudes F, Laroche A, Gaudet D (2007) Anthocyanin expression in marker free transgenic wheat and triticale embryos. In Vitro Cell Dev Biol Plant 43:429–435
Erkens T, Van PM, Vandesompele J, Goossens K, Van ZA, Peelman LJ (2006) Development of a new set of reference genes for normalization of real-time RT-PCR data of porcine backfat and longissimus dorsi muscle, and evaluation with PPARGC1A. BMC Biotechnol 6:41
Fang CX, Xiong J, Qiu L, Wang HB, Song BQ, He HB, Lin RY, Lin WX (2009) Analysis of gene expressions associated with increased allelopathy in rice (Oryza sativa L.) induced by exogenous salicylic acid. Plant Growth Regul 57:163–172
Gachon C, Mingam A, Charrier B (2004) Real-time PCR: what relevance to plant studies? J Exp Bot 55:1445–1454
Ginzinger DG (2002) Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp Hematol 30:503–512
Giulietti A, Overbergh L, Valckx D, Decallonne B, Bouillon R, Mathieu C (2001) An overview of real-time quantitative PCR: applications to quantify cytokine gene expression. Methods 25:386–401
Huggett J, Dheda K, Bustin S, Zumla A (2005) Real-time RT-PCR normalisation; strategies and considerations. Genes Immun 6(4):279–284
Ishitani R, Sunaga K, Hirano A, Saunders P, Katsube N, Chang DM (1996) Evidence that glyceraldehydes-3-phosphate dehydrogenase is involved in age-induced apoptosis in mature cerebellar neurons in culture. J Neurochem 66:928–935
Jarošova J, Kundu J (2010) Validation of reference genes as internal control for studying viral infections in cereals by quantitative real-time RT-PCR. BMC Plant Biol 10:146–154
Kim DH, Kang JG, Kim B (2007) Isolation and characterization of the cytoplasmic male sterility-associated orf456 gene of chill pepper (Capsicum annuum L.). Plant Mol Biol 63:519–532
Lland H, Hertzberg M, Marlton P (2006) Myeloid leukemia. In: Colgan SP (ed) Methods and protocols. Humana, Totowa, NJ, p 53
Narasimha RN, Harinath D, Yamini KN, Sujatha M, Dinesh KV (2009) Expression of sunflower cytoplasmic male sterility-associated open reading frame, orfH522 induces male sterility in transgenic tobacco plants. Planta 229:987–1001
Piston F, Dorado G, Martin A, Barro F (2004) Cloning and characterization of a gamma-3 hordein mRNA (cDNA) from Hordeum chilense. Theor Appl Genet 108:1359–1365
Remans T, Smeets K, Opdenakker K, Mathijsen D, Vangronsveld J, Cuypers A (2008) Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 227:1343–1349
Schmittgen TD, Zakrajsek BA (2000) Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods 46:69–81
Singh R, Green MR (1993) Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science 259:365–368
Sturzenbaum SR, Kille P (2001) Control genes in quantitative molecular biological techniques: the variability of invariance. Comp Biochem Physiol B Biochem Mol Biol 130:281–289
Thellin O, Zorzi W, Lakaye B, Borman DB, Coumans B, Hennen G, Grisar T, Igout A, Heinen E (1999) Housekeeping genes as internal standards: use and limits. J Biotechnol 75:291–295
Vandesompele J, De PK, Pattyn F, Poppe B, Van RN, De PA, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3(7): research0034.1-0034.11
Weis JH, Tan SS, Martin BK, Wittwe CT (1992) Detection of rare mRNAs via quantitative RT-PCR. Trends Genet 8:263–264
Yoo WG, Kim TI, Li S, Kwon OS, Cho PY, Kim T, Kim K, Hong S (2009) Reference genes for quantitative analysis on Clonorchis sinensis gene expression by real-time PCR. Parasitol Res 104:321–328
Yusuke I, Koji K, Kyonoshin M, Teruaki T, Masatomo K, Motoaki S, Kazuo S, Kazuo Y (2006) Functional analysis of rice DREB/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol 47(1):141–153
Zhao HL, Ding LP, Sun JB, Wang SB (1995) Selection and identification of CMS lines 21A, 8A and 17A in pepper. Jiangsu Agric Sci 1:49–51
Acknowledgments
This research was supported by the General Program from the Natural Science Foundation of Jiangsu (BK2010464); National Staple Vegetables Industrial Technology System Huai’an Experiment Station Project; Jiangsu Programs CX (11)104 and CX (10)103.
Author information
Authors and Affiliations
Corresponding author
Additional information
Wang Shu Bin and Liu Ke Wei have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bin, W.S., Wei, L.K., Ping, D.W. et al. Evaluation of appropriate reference genes for gene expression studies in pepper by quantitative real-time PCR. Mol Breeding 30, 1393–1400 (2012). https://doi.org/10.1007/s11032-012-9726-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-012-9726-7