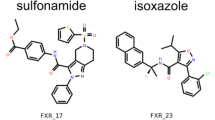
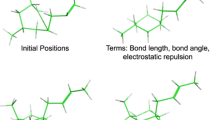
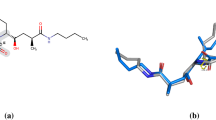
Abstract
We describe a new template-based method for docking flexible ligands such as macrocycles to proteins. It combines Monte-Carlo energy minimization on the manifold, a fast manifold search method, with BRIKARD for complex flexible ligand searching, and with the MELD accelerator of Replica-Exchange Molecular Dynamics simulations for atomistic degrees of freedom. Here we test the method in the Drug Design Data Resource blind Grand Challenge competition. This method was among the best performers in the competition, giving sub-angstrom prediction quality for the majority of the targets.











Similar content being viewed by others
References
Padhorny D, Hall DR, Mirzaei H et al (2018) Protein–ligand docking using FFT based sampling: D3R case study. J Comput Aided Mol Des 32:225–230. https://doi.org/10.1007/s10822-017-0069-7
Ignatov M, Liu C, Alekseenko A et al (2019) Monte Carlo on the manifold and MD refinement for binding pose prediction of protein–ligand complexes: 2017 D3R Grand Challenge. J Comput Aided Mol Des 33:119–127. https://doi.org/10.1007/s10822-018-0176-0
Vassar R, Bennett BD, Babu-Khan S et al (1999) Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science 286:735–741. https://doi.org/10.1126/science.286.5440.735
Coutsias EA, Lexa KW, Wester MJ et al (2016) Exhaustive conformational sampling of complex fused ring macrocycles using inverse kinematics. J Chem Theory Comput 12:4674–4687. https://doi.org/10.1021/acs.jctc.6b00250
Mirzaei H, Beglov D, Paschalidis IC et al (2012) Rigid body energy minimization on manifolds for molecular docking. J Chem Theory Comput 8:4374–4380. https://doi.org/10.1021/ct300272j
Mirzaei H, Zarbafian S, Villar E et al (2015) Energy minimization on manifolds for docking flexible molecules. J Chem Theory Comput 11:1063–1076. https://doi.org/10.1021/ct500155t
Perez A, MacCallum JL, Dill KA (2015) Accelerating molecular simulations of proteins using Bayesian inference on weak information. Proc Natl Acad Sci USA 112:11846–11851. https://doi.org/10.1073/pnas.1515561112
MacCallum JL, Perez A, Dill KA (2015) Determining protein structures by combining semireliable data with atomistic physical models by Bayesian inference. Proc Natl Acad Sci USA 112:6985–6990. https://doi.org/10.1073/pnas.1506788112
Riniker S, Landrum GA (2015) Better informed distance geometry: using what we know to improve conformation generation. J Chem Inf Model 55:2562–2574. https://doi.org/10.1021/acs.jcim.5b00654
Landrum G (2019) RDKit: Open-source cheminformatics. https://rdkit.org/. Accessed 10 July 2019
Coutsias EA, Wester MJ (2019) Multiple loop closure and ring perception (unpublished)
Halgren TA (1996) Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem 17:490–519
Tosco P, Stiefl N, Landrum G (2014) Bringing the MMFF force field to the RDKit: implementation and validation. J Cheminform 6:37. https://doi.org/10.1186/s13321-014-0037-3
Moghadasi M, Mirzaei H, Mamonov A et al (2015) The impact of side-chain packing on protein docking refinement. J Chem Inf Model 55:872–881. https://doi.org/10.1021/ci500380a
Kortemme T, Morozov AV, Baker D (2003) An orientation-dependent hydrogen bonding potential improves prediction of specificity and structure for proteins and protein-protein complexes. J Mol Biol 326:1239–1259. https://doi.org/10.1016/S0022-2836(03)00021-4
Wedemeyer WJ, Baker D (2003) Efficient minimization of angle-dependent potentials for polypeptides in internal coordinates. Proteins 53:262–272. https://doi.org/10.1002/prot.10525
Liu DC, Nocedal J (1989) On the limited memory BFGS method for large scale optimization. Math Program 45:503–528. https://doi.org/10.1007/BF01589116
Mamonov AB, Moghadasi M, Mirzaei H et al (2016) Focused grid-based resampling for protein docking and mapping. J Comput Chem 37:961–970. https://doi.org/10.1002/jcc.24273
Maier JA, Martinez C, Kasavajhala K et al (2015) ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput 11:3696–3713. https://doi.org/10.1021/acs.jctc.5b00255
Wang J, Wolf RM, Caldwell JW et al (2004) Development and testing of a general Amber force field. J Comput Chem 25:1157–1174. https://doi.org/10.1002/jcc.20035
Jakalian A, Bush BL, Jack DB, Bayly CI (2000) Fast, efficient generation of high-quality atomic charges. AM1-BCC model: I. Method. J Comput Chem 21:132–146
Case DA, Betz RM, Cerutti DS et al (2016) AMBER 2016 reference manual. University of California, San Francisco, pp 1–923
Jorgensen WL, Chandrasekhar J, Madura JD et al (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935. https://doi.org/10.1063/1.445869
Joung IS, Cheatham TE 3rd (2008) Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J Phys Chem B 112:9020–9041. https://doi.org/10.1021/jp8001614
Hornak V, Okur A, Rizzo RC, Simmerling C (2006) HIV-1 protease flaps spontaneously open and reclose in molecular dynamics simulations. Proc Natl Acad Sci USA 103:915–920. https://doi.org/10.1073/pnas.0508452103
Wang L, Friesner RA, Berne BJ (2011) Replica exchange with solute scaling: a more efficient version of replica exchange with solute tempering (REST2). J Phys Chem B 115:9431–9438. https://doi.org/10.1021/jp204407d
Ester M, Kriegel H-P, Sander J, Xu X (1996) A density-based algorithm for discovering clusters in large spatial databases with noise. Proceedings of the Second international conference on knowledge discovery and data mining. AAAI Press, Portland, pp 226–231
Pedregosa F, Varoquaux G, Gramfort A et al (2011) Scikit-learn: machine learning in Python. J Mach Learn Res 12:2825–2830
Butina D (1999) Unsupervised data base clustering based on daylight’s fingerprint and tanimoto similarity: a fast and automated way to cluster small and large data sets. J Chem Inf Comput Sci 39:747–750. https://doi.org/10.1021/ci9803381
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. https://doi.org/10.1002/jcc.21334
Duan R, Xu X, Zou X (2018) Lessons learned from participating in D3R 2016 Grand Challenge 2: compounds targeting the farnesoid X receptor. J Comput Aided Mol Des 32:103–111. https://doi.org/10.1007/s10822-017-0082-x
da Silva Figueiredo Celestino GomesGomes P, Da Silva F, Bret G, Rognan D (2018) Ranking docking poses by graph matching of protein-ligand interactions: lessons learned from the D3R Grand Challenge 2. J Comput Aided Mol Des 32:75–87. https://doi.org/10.1007/s10822-017-0046-1
Fradera X, Verras A, Hu Y et al (2018) Performance of multiple docking and refinement methods in the pose prediction D3R prospective Grand Challenge 2016. J Comput Aided Mol Des 32:113–127. https://doi.org/10.1007/s10822-017-0053-2
Lensink MF, Velankar S, Kryshtafovych A et al (2016) Prediction of homoprotein and heteroprotein complexes by protein docking and template-based modeling: a CASP-CAPRI experiment. Proteins 84(Suppl 1):323–348. https://doi.org/10.1002/prot.25007
Kundrotas PJ, Anishchenko I, Badal VD et al (2018) Modeling CAPRI targets 110-120 by template-based and free docking using contact potential and combined scoring function. Proteins 86(Suppl 1):302–310. https://doi.org/10.1002/prot.25380
Peterson LX, Shin W-H, Kim H, Kihara D (2018) Improved performance in CAPRI round 37 using LZerD docking and template-based modeling with combined scoring functions. Proteins 86(Suppl 1):311–320. https://doi.org/10.1002/prot.25376
Porter KA, Padhorny D, Desta I et al (2019) Template-based modeling by ClusPro in CASP13 and the potential for using co-evolutionary information in docking. Protein: Struct Funct Bioinf. https://doi.org/10.1002/prot.25808
Zheng J, Kundrotas PJ, Vakser IA, Liu S (2016) Template-based modeling of protein-RNA interactions. PLoS Comput Biol 12:e1005120. https://doi.org/10.1371/journal.pcbi.1005120
Zheng J, Hong X, Xie J et al (2019) P3DOCK: a protein-RNA docking webserver based on template-based and template-free docking. Bioinformatics. https://doi.org/10.1093/bioinformatics/btz478
Kadukova M, Grudinin S (2017) Convex-PL: a novel knowledge-based potential for protein-ligand interactions deduced from structural databases using convex optimization. J Comput Aided Mol Des 31:943–958. https://doi.org/10.1007/s10822-017-0068-8
Jiménez J, Škalič M, Martínez-Rosell G, De Fabritiis G (2018) KDEEP: protein-ligand absolute binding affinity prediction via 3D-convolutional neural networks. J Chem Inf Model 58:287–296. https://doi.org/10.1021/acs.jcim.7b00650
Nguyen DD, Cang Z, Wu K et al (2019) Mathematical deep learning for pose and binding affinity prediction and ranking in D3R Grand Challenges. J Comput Aided Mol Des 33:71–82. https://doi.org/10.1007/s10822-018-0146-6
Sunseri J, King JE, Francoeur PG, Koes DR (2019) Convolutional neural network scoring and minimization in the D3R 2017 community challenge. J Comput Aided Mol Des 33:19–34. https://doi.org/10.1007/s10822-018-0133-y
Acknowledgements
This work was supported by National Institutes of Health grants R21 GM127952 and 1R01GM125813-01; National Science Foundation grants AF 1816314, AF 1645512, and DBI 1759277; and the National Science Foundation PRAC Award ACI-1713695. Part of this research has been enabled by the Blue Waters sustained-petascale computing project, which is supported by the National Science Foundation (Awards OCI-0725070 and ACI-1238993) and the state of Illinois. Blue Waters is a joint effort of the University of Illinois at Urbana–Champaign and its National Center for Supercomputing Applications. This work was funded by Division of Computing and Communication Foundations (Grant No. AF 1816314).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kotelnikov, S., Alekseenko, A., Liu, C. et al. Sampling and refinement protocols for template-based macrocycle docking: 2018 D3R Grand Challenge 4. J Comput Aided Mol Des 34, 179–189 (2020). https://doi.org/10.1007/s10822-019-00257-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-019-00257-1