Abstract
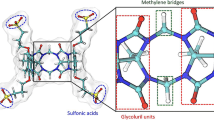
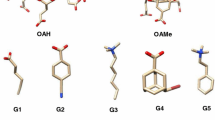
In the context of the SAMPL5 blinded challenge standard free energies of binding were predicted for a dataset of 22 small guest molecules and three different host molecules octa-acids (OAH and OAMe) and a cucurbituril (CBC). Three sets of predictions were submitted, each based on different variations of classical molecular dynamics alchemical free energy calculation protocols based on the double annihilation method. The first model (model A) yields a free energy of binding based on computed free energy changes in solvated and host-guest complex phases; the second (model B) adds long range dispersion corrections to the previous result; the third (model C) uses an additional standard state correction term to account for the use of distance restraints during the molecular dynamics simulations. Model C performs the best in terms of mean unsigned error for all guests (MUE \(3.2\,<\,3.4\,<\,3.6\,\text{kcal}\,\text{mol}^{-1}\)—95 % confidence interval) for the whole data set and in particular for the octa-acid systems (MUE \(1.7\,<\,1.9\,<\,2.1\,\text{kcal}\,\text{mol}^{-1}\)). The overall correlation with experimental data for all models is encouraging (\(R^2\, 0.65\,<\,0.70<0.75\)). The correlation between experimental and computational free energy of binding ranks as one of the highest with respect to other entries in the challenge. Nonetheless the large MUE for the best performing model highlights systematic errors, and submissions from other groups fared better with respect to this metric.





Similar content being viewed by others
Notes
A fourth model that corrects for finite-effects on electrostatic interactions was also submitted by our group, but the results are not discussed here as it was subsequently established that a coding error led to incorrect evaluation of the correction terms.
References
Michel J (2014) Phys Chem Chem Phys 16(10):4465–4477
Geballe MT, Skillman AG, Nicholls A, Guthrie JP, Taylor PJ (2010) J Comput Aided Mol Des 24(4):259–279
Skillman AG (2012) J Comput Aided Mol Des 26(5):473–474
Guthrie JP (2009) J Phys Chem B 113(14):4501–4507
Michel J, Henchman RH, Gerogiokas G, Southey MWY, Mazanetz MP, Law RJ (2014) J Chem Theory Comput 10(9):4055–4068
Jorgensen LW, Thomas LL (2008) J Chem Theory Comput 4(6):869–876
Woods CJ, Malaisree M, Hannongbua S, Mulholland AJ (2011) J Chem Phys. doi:10.1063/1.3519057
Chang C-E, Gilson MK (2004) J Am Chem Soc 126(40):13156–13164
Muddana SH, Gilson MK (2012) J Chem Theory Comput 8(6):2023–2033
Mikulskis P, Cioloboc D, Andrejić M, Khare S, Brorsson J, Genheden S, Mata RA, Söderhjelm P, Ryde U (2014) J Comput Aided Mol Des 28(4):375–400
König G, Pickard IV FC, Mei Y, Brooks BR (2014) J Comput Aided Mol Des 28(3):245–257
Beckstein O, Fourrier A, Iorga BI (2014) J Comput Aided Mol Des 28(3):265–276
Monroe JI, Shirts MR (2014) J Comput Aided Mol Des 28(4):401–415
Chen I-J, Foloppe N (2011) Drug Develop Res 72(1):85–94
Halgren TA, Damm W (2001) Curr Opin Struct Biol 11(2):236–242
Kastenholz MA, Hnenberger PH (2004) J Phys Chem B 108(2):774–788
Jorgensen LW, Ravimohan C (1985) J Chem Phys 83(6):3050–3054
Mezei M (1987) J Chem Phys 86(12):7084–7088
Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE (2000) Acc Chem Res 33(12):889–897
Gibb CLD, Gibb CB (2013) J Comput Aided Mol Des 28(4):319–325
Gilberg L, Zhang B, Zavalij PY, Sindelar V, Isaacs L (2015) Org Biomol Chem 13:4041–4050
Zhang B, Isaacs L (2014) J Med Chem 57(22):9554–9563
Sure R, Antony J, Grimme S (2014) J Phys Chem B 118(12):3431–3440
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) J Comput Chem 25(9):1157–1174
Mishra SK, Calabr G, Loeffler HH, Michel J, Koa J (2015) J Chem Theory Comput 11(7):3333–3345
Aldeghi M, Heifetz A, Bodkin MJ, Knapp S, Biggin PC (2016) Chem Sci 7(1):207–218
Jorgensen WL, Buckner JK, Boudon S, Tirado-Rives J (1988) J Chem Phys 89(6):3742–3746
Gilson MK, Given JA, Bush BL, McCammon JA (1997) Biophys J 72(3):1047
Michel J, Essex JW (2010) J Comput Aided Mol Des 24(8):639–658
Shirts MR, Mobley DL, Chodera JD, Pande VS (2007) J Phys Chem B 111(45):13052–13063
Zwanzig WR (1954) J Chem Phys 22(8):1420–1426
Frenkel D, Smit B (2001) Understanding molecular simulation, 2nd edn. Academic Press Inc, Orlando
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) J Chem Phys 79(2):926–935
Case DA, Babin V, Berryman JT, Betz RM, Cai Q, Cerutti DS, Cheatham III DS, Darden TA, Duke TA, Gohlke H, Goetz AW, Gusarov S, Homeyer N, Janowski P, Kaus J, Kolossvary I, Kovalenko A, Lee TS, LeGrand S, Luchko T, Luo R, Madej B, Merz KM, Paesani F, Roe DR, Roitberg A, Sagui C, Salomon-Ferrer R, Seabra G, Simmerling CL, Smith W, Swails J, Walker RC, Wang J, Wolf RM, Wu X, Kollman PA (2014) AMBER 14, University of California, San Francisco
Woods C, Mey ASJ, Calabro G, Michel J (2016) Sire molecular simulations framework. http://siremol.org. Accessed May 31
Eastman P, Friedrichs MS, Chodera JD, Radmer RJ, Bruns CM, Ku JP, Beauchamp KA, Lane TJ, Wang L-P, Shukla D, Tye T, Houston M, Stich T, Klein C, Shirts MR, Pande VS (2013) J Chem Theory Comput 9(1):461–469
Roe RD, Cheatham TE III (2013) J Chem Theory Comput 9(7):3084–3095
Muddana HS, Fenley AT, Mobley DL, Gilson MK (2014) J Comput Aided Mol Des 28(4):305–317
Schrödinger release 2015-2: Maestro, version 10.2, schrödinger, llc, New York, NY, 2015
Jakalian A, Bush BL, Jack DB, Bayly CI (2000) J Comput Chem 21(2):132–146 (cited By 552)
Shirts MR, Chodera JD (2008) J Chem Phys. doi:10.1063/1.2978177
Hopkins CW, Le Grand S, Walker RC, Roitberg AE (2015) J Chem Theory Comput 11(4):1864–1874
Andersen HC (1980) J Chem Phys 72:2384–2393
Tironi IG, Sperb R, Smith PE, van Gunsteren WF (1995) J Chem Phys 102(13):5451–5459
Gan H, Benjamin CJ, Gibb BC (2011) J Am Chem Soc 133(13):4770–4773
Yin J, Henriksen NM, Slochower DR, Chiu MW, Mobley DL, Gilson MK (2016) Overview of the SAMPL5 host-guest challenge: are we doing better? J Comput Aided Mol Des (under review)
Bosisio S, Mey ASJS, Michel J (2016) Blinded predictions of distribution coefficients in the SAMPL5 challenge. J Comput Aided Mol Des (under review)
Chemaxon. www.chemicalize.org
Yin J, Henriksen NM, Slochower DR, Gilson MK (2016) The SAMPL5 host-guest challenge: binding free energies and enthalpies from explicit solvent simulations. J Comput Aided Mol Des (under review)
Goetz AW, Poole D, Le Grand S, Walker RC, Salomon-Ferrer R (2013) J Chem Theory Comput 9:3878–3888
Velez-Vega C, Gilso MK (2013) J Comput Chem 34(27):2360–2371
Kastenholz MA, Hünenberger PH (2006) J Chem Phys 124(12):124106
Kastenholz MA, Hünenberger PH (2006) J Chem Phys 124(22):224501
Reif MM, Oostenbrink C (2014) J Comput Chem 35(3):227–243
Rocklin GJ, Boyce SE, Fischer M, Fish I, Mobley DL, Shoichet BK, Dill KA (2013) J Mol Biol 425(22):4569–4583
Acknowledgments
J. M. is supported by a Royal Society University Research Fellowship. The research leading to these results has received funding from the European Research Council under the European Unions Seventh Framework Programme (FP7/ 2007-2013)/ERC Grant Agreement No. 336289.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bosisio, S., Mey, A.S.J.S. & Michel, J. Blinded predictions of host-guest standard free energies of binding in the SAMPL5 challenge. J Comput Aided Mol Des 31, 61–70 (2017). https://doi.org/10.1007/s10822-016-9933-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-016-9933-0