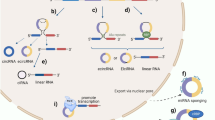
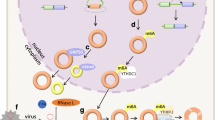
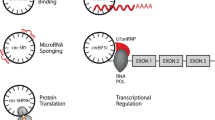
Abstract
Circular RNAs (circRNAs) are a type of single-stranded RNA molecules that normally do not encode proteins. circRNAs are involved in many physiological processes as well as the pathogenesis of diseases. Cardiac fibrosis is increasingly recognized as a pathological force in advanced heart diseases. A growing number of studies have reported that the occurrence and development of cardiac fibrosis is closely associated with the regulation of circRNAs. This review summarizes the current understanding of circRNA biogenesis and function and will highlight the recent updates regarding the involvement of circRNAs in cardiac fibrosis, and their potential as emerging biomarkers and therapeutic targets.



Similar content being viewed by others
References
de Mestral C, Stringhini S (2017) Socioeconomic status and cardiovascular disease: an update. Curr Cardiol Rep 19(11):115
Roth GA, Johnson C, Abajobir A, Abd-Allah F, Abera SF, Abyu G, Ahmed M, Aksut B, Alam T, Alam K, Alla F, Alvis-Guzman N, Amrock S, Ansari H, Ärnlöv J, Asayesh H, Atey TM, Avila-Burgos L, Awasthi A, Banerjee A, Barac A, Bärnighausen T, Barregard L, Bedi N, Belay Ketema E, Bennett D, Berhe G, Bhutta Z, Bitew S, Carapetis J, Carrero JJ, Malta DC, Castañeda-Orjuela CA, Castillo-Rivas J, Catalá-López F, Choi JY, Christensen H, Cirillo M, Cooper L Jr, Criqui M, Cundiff D, Damasceno A, Dandona L, Dandona R, Davletov K, Dharmaratne S, Dorairaj P, Dubey M, Ehrenkranz R, el Sayed Zaki M, Faraon EJA, Esteghamati A, Farid T, Farvid M, Feigin V, Ding EL, Fowkes G, Gebrehiwot T, Gillum R, Gold A, Gona P, Gupta R, Habtewold TD, Hafezi-Nejad N, Hailu T, Hailu GB, Hankey G, Hassen HY, Abate KH, Havmoeller R, Hay SI, Horino M, Hotez PJ, Jacobsen K, James S, Javanbakht M, Jeemon P, John D, Jonas J, Kalkonde Y, Karimkhani C, Kasaeian A, Khader Y, Khan A, Khang YH, Khera S, Khoja AT, Khubchandani J, Kim D, Kolte D, Kosen S, Krohn KJ, Kumar GA, Kwan GF, Lal DK, Larsson A, Linn S, Lopez A, Lotufo PA, el Razek HMA, Malekzadeh R, Mazidi M, Meier T, Meles KG, Mensah G, Meretoja A, Mezgebe H, Miller T, Mirrakhimov E, Mohammed S, Moran AE, Musa KI, Narula J, Neal B, Ngalesoni F, Nguyen G, Obermeyer CM, Owolabi M, Patton G, Pedro J, Qato D, Qorbani M, Rahimi K, Rai RK, Rawaf S, Ribeiro A, Safiri S, Salomon JA, Santos I, Santric Milicevic M, Sartorius B, Schutte A, Sepanlou S, Shaikh MA, Shin MJ, Shishehbor M, Shore H, Silva DAS, Sobngwi E, Stranges S, Swaminathan S, Tabarés-Seisdedos R, Tadele Atnafu N, Tesfay F, Thakur JS, Thrift A, Topor-Madry R, Truelsen T, Tyrovolas S, Ukwaja KN, Uthman O, Vasankari T, Vlassov V, Vollset SE, Wakayo T, Watkins D, Weintraub R, Werdecker A, Westerman R, Wiysonge CS, Wolfe C, Workicho A, Xu G, Yano Y, Yip P, Yonemoto N, Younis M, Yu C, Vos T, Naghavi M, Murray C (2017) Global, regional, and national burden of cardiovascular diseases for 10 causes, 1990 to 2015. J Am Coll Cardiol 70(1):1–25
Fan D, Takawale A, Lee J, Kassiri Z (2012) Cardiac fibroblasts, fibrosis and extracellular matrix remodeling in heart disease. Fibrogenesis Tissue Repair 5(1):15
Kong P, Christia P, Frangogiannis NG (2014) The pathogenesis of cardiac fibrosis. Cell Mol Life Sci 71(4):549–574
Talman V, Ruskoaho H (2016) Cardiac fibrosis in myocardial infarction—from repair and remodeling to regeneration. Cell Tissue Res 365(3):563–581
Krenning G, Zeisberg EM, Kalluri R (2010) The origin of fibroblasts and mechanism of cardiac fibrosis. J Cell Physiol 225(3):631–637
Travers JG, Kamal FA, Robbins J, Yutzey KE, Blaxall BC (2016) Cardiac fibrosis: the fibroblast awakens. Circ Res 118(6):1021–1040
Baicu CF, Stroud JD, Livesay VA, Hapke E, Holder J, Spinale FG et al (2003) Changes in extracellular collagen matrix alter myocardial systolic performance. Am J Phys Heart Circ Phys 284(1):H122–HH32
Briasoulis A, Mallikethi-Reddy S, Palla M, Alesh I, Afonso L (2015) Myocardial fibrosis on cardiac magnetic resonance and cardiac outcomes in hypertrophic cardiomyopathy: a meta-analysis. Heart 101(17):1406–1411
Wapinski O, Chang HY (2011) Long noncoding RNAs and human disease. Trends Cell Biol 21(6):354–361
Barbagallo D, Vittone G, Romani M, Purrello M (2018) Noncoding RNAs in health and disease. Int J Genomics 2018. https://doi.org/10.1155/2018/9135073
Esteller M (2011) Non-coding RNAs in human disease. Nat Rev Genet 12(12):861–874
Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS (2010) Non-coding RNAs: regulators of disease. J Pathol 220(2):126–139
Beermann J, Piccoli M-T, Viereck J, Thum T (2016) Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev 96(4):1297–1325
Devaux Y, Zangrando J, Schroen B, Creemers EE, Pedrazzini T, Chang C-P, Dorn GW 2nd, Thum T, Heymans S, Cardiolinc network (2015) Long noncoding RNAs in cardiac development and ageing. Nat Rev Cardiol 12(7):415–425
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297
Fan X, Weng X, Zhao Y, Chen W, Gan T, Xu D (2017) Circular RNAs in cardiovascular disease: an overview. Biomed Res Int 2017
Fischer JW, Leung AK (2017) CircRNAs: a regulator of cellular stress. Crit Rev Biochem Mol Biol 52(2):220–233
Panda AC, Grammatikakis I, Munk R, Gorospe M, Abdelmohsen K (2017) Emerging roles and context of circular RNAs. WIREs 8(2):e1386
Chen L-L (2016) The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 17(4):205–211
Du WW, Zhang C, Yang W, Yong T, Awan FM, Yang BB (2017) Identifying and characterizing circRNA-protein interaction. Theranostics 7(17):4183–4191
Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P et al (1993) Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73(5):1019–1030
Cocquerelle C, Daubersies P, Majerus M-A, Kerckaert J-P, Bailleul B (1992) Splicing with inverted order of exons occurs proximal to large introns. EMBO J 11(3):1095–1098
Zaphiropoulos PG (1996) Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci 93(13):6536–6541
Zeng X, Lin W, Guo M, Zou Q. A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput Biol 2017;13(6):\
Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna 19(2):141–157
Zhang Y, Zhang X-O, Chen T, Xiang J-F, Yin Q-F, Xing Y-H et al (2013) Circular intronic long noncoding RNAs. Mol Cell 51(6):792–806
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G (2015) Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22(3):256–264
Proudfoot NJ, Furger A, Dye MJ (2002) Integrating mRNA processing with transcription. Cell 108(4):501–512
Burset M, Seledtsov I, Solovyev V (2000) Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res 28(21):4364–4375
Jurica MS, Moore MJ (2003) Pre-mRNA splicing: awash in a sea of proteins. Mol Cell 12(1):5–14
Li X, Yang L, Chen L-L. The biogenesis, functions, and challenges of circular RNAs. Mol Cell 2018
Lasda E, Parker R (2014) Circular RNAs: diversity of form and function. Rna 20(12):1829–1842
Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO (2013) Cell-type specific features of circular RNA expression. PLoS Genet 9(9):e1003777
Jeck WR, Sharpless NE (2014) Detecting and characterizing circular RNAs. Nat Biotechnol 32(5):453
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 495(7441):333–338
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495(7441):384–388
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S (2014) circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56(1):55–66
Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li X et al (2017) A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics 7(16):3842–3855
Kulcheski FR, Christoff AP, Margis R (2016) Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol 238:42–51
Guo JU, Agarwal V, Guo H, Bartel DP (2014) Expanded identification and characterization of mammalian circular RNAs. Genome Biol 15(7):409
Chen G, Shi Y, Liu M, Sun J (2018) circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis 9(2):175
Kai D, Yannian L, Yitian C, Dinghao G, Xin Z, Wu J (2018) Circular RNA HIPK3 promotes gallbladder cancer cell growth by sponging microRNA-124. Biochem Biophys Res Commun 503(2):863–869
Wang J, Zhao S, Ouyang S, Huang Z, Luo Q, Liao L. Circular RNA circHIPK3 acts as the sponge of microRNA-124 to promote human oral squamous cell carcinoma cells proliferation. Zhonghua kou qiang yi xue za zhi 2018;53(8):546–551
Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O et al (2017) Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell 66(1):22–37 e9
Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L et al (2017) Translation of circRNAs. Mol Cell 66(1):9–21 e7
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, Wong CC, Xiao X, Wang Z (2017) Extensive translation of circular RNAs driven by N 6-methyladenosine. Cell Res 27(5):626–641
Han D, Li J, Wang H, Su X, Hou J, Gu Y, Qian C, Lin Y, Liu X, Huang M, Li N, Zhou W, Yu Y, Cao X (2017) Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology 66(4):1151–1164
Hansen TB, Kjems J, Damgaard CK (2013) Circular RNA and miR-7 in cancer. Cancer Res 73(18):5609–5612
Lukiw W (2013) Circular RNA (circRNA) in Alzheimer’s disease (AD). Front Genet 4:307
Venø MT, Hansen TB, Venø ST, Clausen BH, Grebing M, Finsen B et al (2015) Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol 16(1):245
Bayoumi AS, Aonuma T, Teoh J-p, Tang Y-l, Kim I-m (2018) Circular noncoding RNAs as potential therapies and circulating biomarkers for cardiovascular diseases. Acta Pharmacol Sin 39(7):1100
Altesha MA, Ni T, Khan A, Liu K, Zheng X (2019) Circular RNA in cardiovascular disease. J Cell Physiol 234(5):5588–5600
Porter KE, Turner NA (2009) Cardiac fibroblasts: at the heart of myocardial remodeling. Pharmacol Ther 123(2):255–278
Tan AY, Zimetbaum P (2011) Atrial fibrillation and atrial fibrosis. J Cardiovasc Pharmacol 57(6):625–629
Wu Q-Q, Xiao Y, Yuan Y, Ma Z-G, Liao H-H, Liu C et al (2017) Mechanisms contributing to cardiac remodelling. Clin Sci 131(18):2319–2345
Fliss H, Gattinger D (1996) Apoptosis in ischemic and reperfused rat myocardium. Circ Res 79(5):949–956
Scarabelli T, Stephanou A, Rayment N, Pasini E, Comini L, Curello S, Ferrari R, Knight R, Latchman D (2001) Apoptosis of endothelial cells precedes myocyte cell apoptosis in ischemia/reperfusion injury. Circulation 104(3):253–256
Santiago JJ, Dangerfield AL, Rattan SG, Bathe KL, Cunnington RH, Raizman JE, Bedosky KM, Freed DH, Kardami E, Dixon IM (2010) Cardiac fibroblast to myofibroblast differentiation in vivo and in vitro: expression of focal adhesion components in neonatal and adult rat ventricular myofibroblasts. Dev Dyn 239(6):1573–1584
Wang J, Chen H, Seth A, McCulloch CA (2003) Mechanical force regulation of myofibroblast differentiation in cardiac fibroblasts. Am J Phys Heart Circ Phys 285(5):H1871–H1H81
Eyden B (2008) The myofibroblast: phenotypic characterization as a prerequisite to understanding its functions in translational medicine. J Cell Mol Med 12(1):22–37
Burlew BS, Weber KT (2002) Cardiac fibrosis as a cause of diastolic dysfunction. Herz 27(2):92–98
Guo Y, Luo F, Liu Q, Xu D (2017) Regulatory non-coding RNAs in acute myocardial infarction. J Cell Mol Med 21(5):1013–1023
Thum T (2014) Noncoding RNAs and myocardial fibrosis. Nat Rev Cardiol 11(11):655–663
Creemers EE, van Rooij E (2016) Function and therapeutic potential of noncoding RNAs in cardiac fibrosis. Circ Res 118(1):108–118
Wilusz JE, Sunwoo H, Spector DL (2009) Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 23(13):1494–1504
Anastasiadou E, Jacob LS, Slack FJ (2018) Non-coding RNA networks in cancer. Nat Rev Cancer 18(1):5
Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Öhman M, Refojo D, Kadener S, Rajewsky N (2015) Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58(5):870–885
Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC (2014) Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep 9(5):1966–1980
You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, Wang X, Hou J, Liu H, Sun W, Sambandan S, Chen T, Schuman EM, Chen W (2015) Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 18(4):603–610
Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A (2006) Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res 34(8):e63-e
Zhou B, Yu J-W (2017) A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-β1. Biochem Biophys Res Commun 487(4):769–775
Tang C-M, Zhang M, Huang L, Hu Z-q, Zhu J-N, Xiao Z et al (2017) CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci Rep 7:40342
Li M, Ding W, Tariq MA, Chang W, Zhang X, Xu W, Hou L, Wang Y, Wang J (2018) A circular transcript of ncx1 gene mediates ischemic myocardial injury by targeting miR-133a-3p. Theranostics 8(21):5855–5869
Lim TB, Aliwarga E, Luu TDA, Li YP, Ng SL, Annadoray L et al (2019) Targeting the highly abundant circular RNA circSlc8a1 in cardiomyocytes attenuates pressure overload induced hypertrophy. Cardiovasc Res
Sun Y, Yang Z, Zheng B, Zhang X-h, Zhang M-l, Zhao X-s et al (2017) A novel regulatory mechanism of smooth muscle α-actin expression by NRG-1/circACTA2/miR-548f-5p axis. Circ Res 121(6):628–635
Ni H, Li W, Zhuge Y, Xu S, Wang Y, Chen Y et al (2019) Inhibition of circHIPK3 prevents angiotensin II-induced cardiac fibrosis by sponging miR-29b-3p. Int J Cardiol
Zhu Y, Pan W, Yang T, Jiang Z, Meng X, Tao L et al (2019) Upregulation of circular RNA circNFIB attenuates cardiac fibrosis by sponging miR-433. Front Genet 10:564
Huang S, Li X, Zheng H, Si X, Li B, Wei G, et al. Loss of super-enhancer-regulated CircRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation. 2019
Garikipati VNS, Verma SK, Cheng Z, Liang D, Truongcao MM, Cimini M et al (2019) Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat Commun 10(1):1–14
Geng H-H, Li R, Su Y-M, Xiao J, Pan M, Cai X-X, Ji XP (2016) The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS One 11(3):e0151753
Wang K, Long B, Liu F, Wang J-X, Liu C-Y, Zhao B, Zhou LY, Sun T, Wang M, Yu T, Gong Y, Liu J, Dong YH, Li N, Li PF (2016) A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 37(33):2602–2611
Cai L, Qi B, Wu X, Peng S, Zhou G, Wei Y, Xu J, Chen S, Liu S (2019) Circular RNA Ttc3 regulates cardiac function after myocardial infarction by sponging miR-15b. J Mol Cell Cardiol 130:10–22
Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W et al (2016) Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun 7:12429
Du WW, Yang W, Chen Y, Wu Z-K, Foster FS, Yang Z et al (2016) Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J 38(18):1402–1412
Ebbesen KK, Kjems J, Hansen TB (2016) Circular RNAs: identification, biogenesis and function. BBA-GENE REGUL MECH 1859(1):163–168
Vausort M, Salgado-Somoza A, Zhang L, Leszek P, Scholz M, Teren A, Burkhardt R, Thiery J, Wagner DR, Devaux Y (2016) Myocardial infarction-associated circular RNA predicting left ventricular dysfunction. J Am Coll Cardiol 68(11):1247–1248
Li P, Chen S, Chen H, Mo X, Li T, Shao Y et al (2015) Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta 444:132–136
Koh W, Pan W, Gawad C, Fan HC, Kerchner GA, Wyss-Coray T et al (2014) Noninvasive in vivo monitoring of tissue-specific global gene expression in humans. Proc Natl Acad Sci 111(20):7361–7366
Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S (2008) MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 456(7224):980–984
Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Payne JA, Pandya K, Zhang Z, Rosenberg P, Mirotsou M, Dzau VJ (2012) MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res 110(11):1465–1473
Roy S, Khanna S, Hussain S-RA, Biswas S, Azad A, Rink C et al (2009) MicroRNA expression in response to murine myocardial infarction: miR-21 regulates fibroblast metalloprotease-2 via phosphatase and tensin homologue. Cardiovasc Res 82(1):21–29
Stanczyk J, Ospelt C, Karouzakis E, Filer A, Raza K, Kolling C, Gay R, Buckley CD, Tak PP, Gay S, Kyburz D (2011) Altered expression of microRNA-203 in rheumatoid arthritis synovial fibroblasts and its role in fibroblast activation. Arthritis Rheum 63(2):373–381
Abonnenc M, Nabeebaccus AA, Mayr U, Barallobre-Barreiro J, Dong X, Cuello F, Sur S, Drozdov I, Langley SR, Lu R, Stathopoulou K, Didangelos A, Yin X, Zimmermann WH, Shah AM, Zampetaki A, Mayr M (2013) Extracellular matrix secretion by cardiac fibroblasts: role of microRNA-29b and microRNA-30c. Circ Res 113(10):1138–1147
Tsuchiya S, Fujiwara T, Sato F, Shimada Y, Tanaka E, Sakai Y, Shimizu K, Tsujimoto G (2011) MicroRNA-210 regulates cancer cell proliferation through targeting fibroblast growth factor receptor-like 1 (FGFRL1). J Biol Chem 286(1):420–428
Chen S, Puthanveetil P, Feng B, Matkovich SJ, Dorn GW, Chakrabarti S (2014) Cardiac miR-133a overexpression prevents early cardiac fibrosis in diabetes. J Cell Mol Med 18(3):415–421
Duisters RF, Tijsen AJ, Schroen B, Leenders JJ, Lentink V, van der Made I, Herias V, van Leeuwen R, Schellings MW, Barenbrug P, Maessen JG, Heymans S, Pinto YM, Creemers EE (2009) miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res 104(2):170–178
Weiser-Evans MC. Smooth muscle differentiation control comes full circle: the circular noncoding RNA, circActa2, functions as a miRNA sponge to fine-tune α-SMA expression. Am Heart Assoc; 2017
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B et al (2016) Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 7:11215
Hu M, Wei X, Li M, Tao L, Wei L, Zhang M et al (2019) Circular RNA expression profiles of persistent atrial fibrillation in patients with rheumatic heart disease. Anatol J Cardiol 21(1):2
Van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS et al (2008) Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci 105(35):13027–13032
Dawson K, Wakili R, Ördög B, Clauss S, Chen Y, Iwasaki Y et al (2013) MicroRNA29: a mechanistic contributor and potential biomarker in atrial fibrillation. Circulation 127(14):1466–1475
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yousefi, F., Soltani, B.M. Circular RNAs as potential theranostics in the cardiac fibrosis. Heart Fail Rev 26, 195–203 (2021). https://doi.org/10.1007/s10741-019-09908-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10741-019-09908-9