Summary
Bioinformatics screening and molecular docking analyses were utilized to select high affinity peptides targeting translationally controlled tumor protein (TCTP). Selected peptide aptamers were tested towards cancer cell lines with different levels of TCTP expression. One peptide (WGQWPYHC) revealed specific cytotoxicity according to the TCTP expression in tumor cells without affecting normal cells. Western blot analysis showed peptide-induced down-regulation of TCTP as primary target as well as of cell-cycle related downstream proteins (CDK2, CDK6, Cyclin D3) in MOLT-4 leukemia cells. “WGQWPYHC” deserves further analysis for targeted therapy of TCTP-expressing tumor cells.

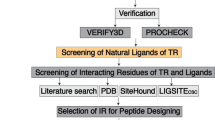
Molecular docking on TCTP, cytotoxicity toward MOLT-4 leukemia cell line and downregulation of CDK2, CDK6, CyclinD3 and TCTP proteins



Similar content being viewed by others
References
Michor F, Nowak MA, Iwasa Y (2006) Evolution of resistance to cancer therapy. Curr Pharm Des 12(3):261–271
Galmarini CM, Galmarini FC (2003) Multidrug resistance in cancer therapy: role of the microenvironment. Curr Opin Investig Drugs 4(12):1416–1421
Tsuruo T, Naito M, Tomida A, Fujita N, Mashima T, Sakamoto H, Haga N (2003) Molecular targeting therapy of cancer: drug resistance, apoptosis and survival signal. Cancer Sci 94(1):15–21
Nishina T, Yamaguchi N, Gohda J, Semba K, Inoue J (2009) NIK is involved in constitutive activation of the alternative NF-kappaB pathway and proliferation of pancreatic cancer cells. Biochem Biophys Res Commun 388(1):96–101. doi:10.1016/j.bbrc.2009.07.125
Huang C, Park CC, Hilsenbeck SG, Ward R, Rimawi MF, Wang YC, Shou J, Bissell MJ, Osborne CK, Schiff R (2011) Beta1 integrin mediates an alternative survival pathway in breast cancer cells resistant to lapatinib. Breast cancer research: BCR 13(4):R84. doi:10.1186/bcr2936
He H, Chen J, Xie WP, Cao S, Hu HY, Yang LQ, Gong B (2013) Ketamine used as an acesodyne in human breast cancer therapy causes an undesirable side effect, upregulating anti-apoptosis protein bcl-2 expression. Genet Mol Res 12(2):1907–1915. doi:10.4238/2013.January.4.7
Voelcker V, Sticherling M (2011) Acneiform skin lesions as a side effect of therapy with EGFR (epidermal growth factor receptor) inhibitors in Colon Cancer. J Dtsch Dermatol Ges 9:220–220
Gamboa EO, Rehmus EH, Haller N (2010) Fournier’s gangrene as a possible side effect of bevacizumab therapy for resected colorectal cancer. Clin Colorectal Canc 9(1):55–60. doi:10.3816/Ccc.2010.N.008
Repetto-Llamazares AHV, Larsen RH, Patzke S, Fleten KG, Didierlaurent D, Pichard A, Pouget JP, Dahle J (2015) Targeted cancer therapy with a novel anti-CD37 Beta-Particle Emitting radioimmunoconjugate for treatment of non-Hodgkin Lymphoma. PLoS One 10(6). doi:10.1371/journal.pone.0128816
Liu H, Lu J, Hua Y, Zhang P, Liang Z, Ruan L, Lian C, Shi H, Chen K, Tu Z (2015) Targeting heat-shock protein 90 with ganetespib for molecularly targeted therapy of gastric cancer. Cell Death Dis 6. doi:10.1038/Cddis.2014.555
Carneiro BA, Altman JK, Kaplan JB, Ossenkoppele G, Swords R, Platanias LC, Giles FJ (2015) Targeted therapy of acute myeloid leukemia. Expert Rev Anticancer Ther 15(4):399–413. doi:10.1586/14737140.2015.1004316
Panathur N, Dalimba U, Koushik PV, Alvala M, Yogeeswari P, Sriram D, Kumar V (2013) Identification and characterization of novel indole based small molecules as anticancer agents through SIRT1 inhibition. Eur J Med Chem 69:125–138. doi:10.1016/j.ejmech.2013.08.018
Gurova K (2009) New hopes from old drugs: revisiting DNA-binding small molecules as anticancer agents. Future Oncol 5(10):1685–1704. doi:10.2217/fon.09.127
Seneci P (2012) Small molecules as pro-apoptotic anticancer agents. Pharm Pat Anal 1(4):483–505. doi:10.4155/ppa.12.41
Mendelsohn J (2003) Antibody-mediated EGF receptor blockade as an anticancer therapy: from the laboratory to the clinic. Cancer Immunol Immunother: CII 52(5):342–346. doi:10.1007/s00262-002-0354-7
Kim DG, Jin Y, Jin J, Yang H, Joo KM, Lee WS, Shim SR, Kim SW, Yoo J, Lee SH, Yoo JS, Nam DH (2015) Anticancer activity of TTAC-0001, a fully human anti-vascular endothelial growth factor receptor 2 (VEGFR-2/KDR) monoclonal antibody, is associated with inhibition of tumor angiogenesis. MAbs 7(6):1195–1204. doi:10.1080/1942s0862.2015.1086854
Kadioglu O, Malczyk AH, Greten HJ, Efferth T (2015) Aptamers as a novel tool for diagnostics and therapy. Invest New Drug 33(2):513–520. doi:10.1007/s10637-015-0213-y
Mayer G (2009) The chemical biology of aptamers. Angew Chem Int Edit 48(15):2672–2689. doi:10.1002/anie.200804643
Hall DA, Ptacek J, Snyder M (2007) Protein microarray technology. Mech Ageing Dev 128(1):161–167. doi:10.1016/j.mad.2006.11.021
Rhinehardt KL, Mohan RV, Srinivas G (2015) Computational modeling of peptide-aptamer binding. Methods Mol Biol 1268:313–333. doi:10.1007/978-1-4939-2285-7_14
Barbas AS, Mi J, Clary BM, White RR (2010) Aptamer applications for targeted cancer therapy. Future Oncol 6(7):1117–1126. doi:10.2217/fon.10.67
Sa LT, Simmons S, Missailidis S, da Silva MI, Santos-Oliveira R (2013) Aptamer-based nanoparticles for cancer targeting. J Drug Target 21(5):427–434. doi:10.3109/1061186X.2012.761222
Westermaier Y, Barril X, Scapozza L (2015) Virtual screening: an in silico tool for interlacing the chemical universe with the proteome. Methods 71:44–57. doi:10.1016/j.ymeth.2014.08.001
Cerqueira NM, Gesto D, Oliveira EF, Santos-Martins D, Bras NF, Sousa SF, Fernandes PA, Ramos MJ (2015) Receptor-based virtual screening protocol for drug discovery. Arch Biochem Biophys 582:56–67. doi:10.1016/j.abb.2015.05.011
Dallakyan S, Olson AJ (2015) Small-molecule library screening by docking with PyRx. Methods Mol Biol 1263:243–250. doi:10.1007/978-1-4939-2269-7_19
Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, Wang J, Yu B, Zhang J, Bryant SH (2015) PubChem substance and compound databases. Nucleic Acids Res. doi:10.1093/nar/gkv951
Ghasemi JB, Shiri F, Pirhadi S, Heidari Z (2015) Discovery of new potential antimalarial compounds using virtual screening of ZINC database. Comb Chem High Throughput Screen 18(2):227–234
Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, Overington JP (2012) ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res 40(Database issue):D1100–D1107. doi:10.1093/nar/gkr777
Duprez W, Bachu P, Stoermer MJ, Tay S, McMahon RM, Fairlie DP, Martin JL (2015) Virtual screening of peptide and peptidomimetic fragments targeted to inhibit bacterial dithiol oxidase DsbA. PLoS One 10(7):e0133805. doi:10.1371/journal.pone.0133805
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461. doi:10.1002/jcc.21334
Park H, Lee J, Lee S (2006) Critical assessment of the automated AutoDock as a new docking tool for virtual screening. Proteins 65(3):549–554. doi:10.1002/prot.21183
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated Docking with selective receptor flexibility. J Comput Chem 30(16):2785–2791. doi:10.1002/Jcc.21256
Morris GM, Huey R, Olson AJ (2008) Using AutoDock for ligand-receptor docking. Current protocols in bioinformatics/editoral board, Andreas D Baxevanis [et al] Chapter 8:Unit 8 14. doi:10.1002/0471250953.bi0814s24
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19(14):1639–1662. doi:10.1002/(Sici)1096-987x(19981115)19:14<1639::Aid-Jcc10>3.0.Co;2-B
Fuhrmann J, Rurainski A, Lenhof HP, Neumann D (2010) A new Lamarckian Genetic Algorithm for Flexible Ligand-Receptor Docking. J Comput Chem 31(9):1911–1918. doi:10.1002/jcc.21478
Wang T (2008) Function and dynamics of aptamers: A case study on the malachite. Ph.D. thesis, Iowa State University, Ames, Iowa
Chushak Y, Stone MO (2009) In silico selection of RNA aptamers. Nucleic Acids Res 37(12). doi:10.1093/nar/gkp408
Aubin-Tam ME, Appleyard DC, Ferrari E, Garbin V, Fadiran OO, Kunkel J, Lang MJ (2011) Adhesion through single peptide aptamers. J Phys Chem A 115(16):3657–3664. doi:10.1021/jp1031493
Edgar RC, Batzoglou S (2006) Multiple sequence alignment. Curr Opin Struc Biol 16(3):368–373. doi:10.1016/j.sbi.2006.04.004
Pei JM (2008) Multiple protein sequence alignment. Curr Opin Struc Biol 18(3):382–386. doi:10.1016/j.sbi.2008.03.007
Tsubery H, Mironchik M, Fridkin M, Shechter Y (2004) Prolonging the action of protein and peptide drugs by a novel approach of reversible polyethylene glycol modification. J Biol Chem 279(37):38118–38124. doi:10.1074/jbc.M405155200
Grun J, Revell JD, Conza M, Wennemers H (2006) Peptide-polyethylene glycol conjugates: synthesis and properties of peptides bearing a C-terminal polyethylene glycol chain. Bioorgan Med Chem 14(18):6197–6201. doi:10.1016/j.bmc.2006.05.079
Acunzo J, Baylot V, So A, Rocchi P (2014) TCTP as therapeutic target in cancers. Cancer Treat Rev 40(6):760–769. doi:10.1016/j.ctrv.2014.02.007
Miao X, Chen YB, Xu SL, Zhao T, Liu JY, Li YR, Wang J, Zhang J, Guo GZ (2013) TCTP overexpression is associated with the development and progression of glioma. Tumor Biol 34(6):3357–3361. doi:10.1007/s13277-013-0906-9
Bommer UA, Thiele BJ (2004) The translationally controlled tumour protein (TCTP). Int J Biochem Cell B 36(3):379–385. doi:10.1016/S1357-2725(03)00213-9
Tuynder M, Fiucci G, Prieur S, Lespagnol A, Geant A, Beaucourt S, Duflaut D, Besse S, Susini L, Cavarelli J, Moras D, Amson R, Telerman A (2004) Translationally controlled tumor protein is a target of tumor reversion. P Natl Acad Sci USA 101(43):15364–15369. doi:10.1073/pnas.0406776101
Lucibello M, Gambacurta A, Zonfrillo M, Pierimarchi P, Serafino A, Rasi G, Rubartelli A, Garaci E (2011) TCTP is a critical survival factor that protects cancer cells from oxidative stress-induced cell-death. Exp Cell Res 317(17):2479–2489. doi:10.1016/j.yexcr.2011.07.012
Tsarova K, Yarmola EG, Bubb MR (2010) Identification of a cofilin-like actin-binding site on translationally controlled tumor protein (TCTP). FEBS Lett 584(23):4756–4760. doi:10.1016/j.febslet.2010.10.054
Yang Y, Yang F, Xiong Z, Yan Y, Wang X, Nishino M, Mirkovic D, Nguyen J, Wang H, Yang XF (2005) An N-terminal region of translationally controlled tumor protein is required for its antiapoptotic activity. Oncogene 24(30):4778–4788. doi:10.1038/sj.onc.1208666
Jung J, Kim M, Kim MJ, Kim J, Moon J, Lim JS, Kim M, Lee K (2004) Translationally controlled tumor protein interacts with the third cytoplasmic domain of Na,K-ATPase alpha subunit and inhibits the pump activity in HeLa cells. J Biol Chem 279(48):49868–49875. doi:10.1074/jbc.M400895200
Rho SB, Lee JH, Park MS, Byun HJ, Kang S, Seo SS, Kim JY, Park SY (2011) Anti-apoptotic protein TCTP controls the stability of the tumor suppressor p53. FEBS Lett 585(1):29–35. doi:10.1016/j.febslet.2010.11.014
Yoon T, Jung J, Kim M, Lee KM, Choi EC, Lee K (2000) Identification of the self-interaction of rat TCTP/IgE-dependent histamine-releasing factor using yeast two-hybrid system. Arch Biochem Biophys 384(2):379–382. doi:10.1006/abbi.2000.2108
Gachet Y, Tournier S, Lee M, Lazaris-Karatzas A, Poulton T, Bommer UA (1999) The growth-related, translationally controlled protein P23 has properties of a tubulin binding protein and associates transiently with microtubules during the cell cycle. J Cell Sci 112(Pt 8):1257–1271
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) Open Babel: An open chemical toolbox. J Cheminf 3:33. doi:10.1186/1758-2946-3-33
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14(1):33–38 27-38
Cui Q, Lim SK, Zhao B, Hoffmann FM (2005) Selective inhibition of TGF-beta responsive genes by smad-interacting peptide aptamers from FoxH1, Lef1 and CBP. Oncogene 24(24):3864–3874. doi:10.1038/sj.onc.1208556
O’Brien J, Wilson I, Orton T, Pognan F (2000) Investigation of the alamar blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur J Biochem/FEBS 267(17):5421–5426
Kuete V, Wiench B, Hegazy ME, Mohamed TA, Fankam AG, Shahat AA, Efferth T (2012) Antibacterial activity and cytotoxicity of selected Egyptian medicinal plants. Planta Med 78(2):193–199. doi:10.1055/s-0031-1280319
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We declare that there is no conflict of interest.
Rights and permissions
About this article
Cite this article
Kadioglu, O., Efferth, T. Peptide aptamer identified by molecular docking targeting translationally controlled tumor protein in leukemia cells. Invest New Drugs 34, 515–521 (2016). https://doi.org/10.1007/s10637-016-0339-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10637-016-0339-6