Abstract
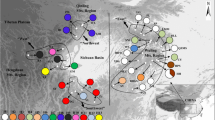
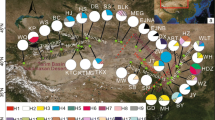
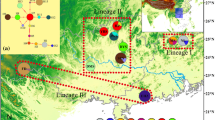
Gymnocarpos przewalskii is restricted mainly to the arid desert areas of northwestern China, and is listed as a rare and endangered species of second conservation priority in the China Red Data Book. A phylogeographic study was conducted using two chloroplast regions (psbA–trnH and ycf6–psbM), together with various population genetic analyses, to examine genetic variation, population structure, and evolutionary history of the species. In all, 25 haplotypes were detected, 14 of which were found in the northwestern Tarim Basin of southern Xinjiang Autonomous Region. G. przewalskii showed high levels of total genetic diversity (h T = 0.849) and average gene diversity within populations (h S = 0.350). A higher N ST than G ST (P < 0.001) indicated significant phylogeographic structure across the species range. Nested clade phylogeographic analysis (NCPA) suggested that genetic structure in G. przewalskii has been heavily affected by past fragmentation, which likely resulted from aridity and the expansion of desert in northwestern China during the Quaternary. Mismatch analyses were consistent with NCPA in additionally suggesting contiguous range expansion for populations in the northern Tarim Basin and the Yumen region of Gansu Province, which may have occurred during a less arid interval. Human disturbance was identified as the greatest current threat to the species.



Similar content being viewed by others
References
Bandelt HJ, Forster P, Röhl A (1999) Median joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Bohonak AJ (2002) IBD (isolation by distance): a program for analyses of isolation by distance. J Hered 93:153–154
Chang ZY, Xu LR, Wu ZH (2004) Comparative morphology, ecology and geographical distribution of Calophaca sinica and C. soongorica. Acta Bot Bor-Occid Sin 24:2312–2320
Chen GQ, Crawford D, Huang HW, Ge XJ (2009a) Genetic structure and mating system of Ammopiptanthus mongolicus (Leguminosae), an endangered shrub in north-western China. Plant Spec Biol 24:179–188
Chen GQ, Huang HW, Crawford DJ, Pan BR, Ge XJ (2009b) Mating system and genetic diversity of a rare desert legume Ammopiptanthus nanus (Leguminosae). J Syst Evol 47:57–66
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Comes HP, Kadereit JW (1998) The effect of Quaternary climatic changes on plant distribution and evolution. Trends Plant Sci 3:432–438
Cox CB, Moore PD (2000) Biogeography: an ecological and evolutionary approach. Blackwell Science, Oxford
Crandall KA, Templeton AR (1993) Empirical tests of some predictions from coalescent theory with applications to intraspecific phylogeny reconstruction. Genetics 134:959–969
Dang RL, Pan XL, Gu XF (2002) Floristic analysis of spermatophyte genera in the arid deserts area in North-West China. Guihaia 22:121–128
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Dupanloup I, Schneider S, Excoffier L (2002) A simulated annealing approach to define the genetic structure of populations. Mol Ecol 11:2571–2581
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: applications to human mitochondrial DNA restriction data. Genetics 131:479–491
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform 1:47–50
Falchi A, Paolini J, Desjobert JM, Melis A, Costa J, Varesi L (2009) Phylogeography of Cistus creticus L. on Corsica and Sardinia inferred by the TRNL-F and RPL32-TRNL sequences of cpDNA. Mol Phylogenet Evol 52:538–543
Fan ZL (1993) A study on the formation and evolution of oases in Tarim Basin. Acta Geographica Sin 5:421–427
Frankel OH, Soulé ME (1981) Conservation and evolution. Cambridge University Press, Cambridge
Fu LG (1992) China plant red data book. Science Press, Beijing
Ge XJ, Yu Y, Yuan YM, Huang HW, Yan C (2005a) Genetic diversity and geographic differentiation in endangered Ammopiptanthus (Leguminosae) populations in desert regions of northwest China as revealed by ISSR analysis. Ann Bot 95:843–851
Ge XJ, Zhou XL, Li ZC, Hsu TW, Schaal BA, Ching TY (2005b) Low genetic diversity and significant population structuring in the relict Amentotaxus argotaenia complex (Taxaceae) base on ISSR fingerprinting. J Plant Res 118:415–422
Ge XJ, Hwang CC, Liu ZH, Huang CC, Huang WH, Huang KH, Wang WK, Chiang TY (2011) Conservation genetics and phylogeography of endangered and endemic shrub Tetraena mongolica (Zygophyllaceae) in Inner Mongolia, China. BMC Genet 12:1–12
Guo ZT, Peng SZ, Hao QZ, Chen XH, Liu TS (1999) Late Tertiary development of aridification in northwestern China: link with the arctic ice-sheet formation and Tibetan uplifts. Quat Sci 6:556–566
Guo ZT, Sun B, Zhang ZS, Peng SZ, Xiao GQ, Ge JY, Hao QZ, Qiao YS, Liang MY, Liu JF, Yin QZ, Wei JJ (2008) A major reorganization of Asian climate by the early Miocene. Clim Past 4:153–174
Guo YP, Zhang R, Chen CY, Zhou DW, Liu JQ (2010) Allopatric divergence and regional range expansion of Juniperus sabina in China. J Syst Evol 48:153–160
Hamilton M (1999) Four primer pairs for the amplification of chloroplast intergenic regions with intraspecific variation. Mol Ecol 8:521–523
Harpending HC (1994) Signature of ancient population-growth in a low-resolution mitochondrial-DNA mismatch distribution. Hum Biol 66:591–600
Harrison TM, Yin A, Ryerson FJ (1998) Orographic evolution of the Himalaya and Tibetan plateau. In: Crowley TJ, Burke KC (eds) Tectonic boundary conditions for climate reconstruction. Oxford University Press, Oxford, pp 39–73
Hewitt GM (1996) Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc 58:247–276
Hewitt GM (2000) The genetic legacy of the quaternary ice ages. Nature 405:907–913
Hewitt GM (2004) Genetic consequences of climatic oscillations in the quaternary. Phil Trans R Soc Lond B 359:183–195
Li WC (1998) The Chinese quaternary vegetation and environment. Science Press, Beijing
Mu GJ (1994) On the age and evolution of the Taklamakan desert. Arid Land Geogr 17:1–9
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Oxelman B, Ahlgren B, Thulin M (2002) Circumscription and phylogenetic relationships of Gymnocarpos (Caryophyllaceae-Paronychioideae). Edinb J Bot 59:221–237
Panchal M, Beaumont MA (2007) The automation and evaluation of nested clade phylogeographic analysis. Evolution 61:1466–1480
Petrusson L, Thulin M (1996) Taxonomy and biogeography of Gymnocarpos (Caryophyllaceae). Edinb J Bot 53:1–26
Pons O, Petit RJ (1996) Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144:1237–1245
Posada D, Crandall KA, Templeton AR (2000) GeoDis: a program for the cladistic nested analysis of the geographical distribution of genetic haplotypes. Mol Ecol 9:487–488
Rieseberg LH, Church SA, Morjan CL (2003) Integration of populations and differentiation of species. New Phytol 161:59–69
Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Schneider S, Excoffier L (1999) Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates very among sites: application to human mitochondrial DNA. Genetics 152:1079–1089
Shaw J, Lickey E, Schilling E, Small R (2007) Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am J Bot 94:275–288
Sun JM (2002) Source regions and formation of the loess sediments on the high mountain regions of northwestern China. Quat Res 58:341–351
Sun JM, Zhang LY, Deng CL, Zhu RX (2008) Evidence for enhanced aridity in the Tarim Basin of China since 5.3 Ma. Quat Sci Rev 27:1012–1023
Templeton AR, Boerwinkle E, Sing CF (1987) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. I. Basic theory and an analysis of alcohol dehydrogenase activity in Drosophila. Genetics 117:343–351
Thompson JDTJ, Gibson PF, Jeanmougin F, Higgins DG (1997) The windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Van der Pijl L (1969) Principles of dispersal in higher plants. Springer, Berlin
Wang ZB, Gao QX, Sun JZ, Ma QL (2009) Study on biological characteristics of rare endangered plant Gymnocarpos Przewalskii. Resour Dev Market 25:481–509
Yang D, Fang XM, Dong GR, Peng ZC, Li JJ (2006) Aeolian deposit evidence for formation and evolution of the Tengger Desert in the north of China since early Pleistocene. Mar Geol Quat Geol 26:93–100
Yuan QJ, Zhang ZY, Peng H, Ge S (2008) Chloroplast phylogeography of Dipentodon (Dipentodontaceae) in southwest China and northern Vietnam. Mol Ecol 17:1054–1065
Zheng GZ, Yue LP, He JF, Wang JX, Zhang YL (2006) Grain-size characteristics of the sediments at Palaeoswamp in Anxi County in downstream of Shule River during Holocene and its paleoclimatic significance. Acta Sedimentologica Sin 24:732–739
Acknowledgments
This work was supported by CAS Important Direction for Knowledge Innovation Project (No. KZCX2-EW-305), and Xinjiang Institute of Ecology and Geogeraphy, CAS. We thank Hong-xiang Zhang, Hong-hu Meng and Xiao-jun Shi for their assistance with field survey and sample collection. We are grateful to Qing-long Geng and Xiao-long Jiang for help in haplotype plotting. We would like to give special thanks Dr. Stewart C. Sanderson for his useful comments and English corrections to this manuscript.
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
Variable nucleotide sites in two chloroplast DNA spacers: psbA-trnH and ycf6-psbM, in twenty-five haplotypes of Gymnocarpos przewalskii.

Rights and permissions
About this article
Cite this article
Ma, S., Zhang, M. Phylogeography and conservation genetics of the relic Gymnocarpos przewalskii (Caryophyllaceae) restricted to northwestern China. Conserv Genet 13, 1531–1541 (2012). https://doi.org/10.1007/s10592-012-0397-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-012-0397-z