Abstract
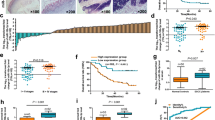
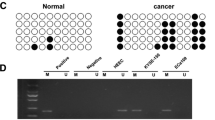
Accumulating evidences indicated that some microRNAs (miRNAs) play a critical role during the carcinogenesis. In the present study, we found that miR-124 is down-regulated in esophageal squamous cell carcinoma (ESCC) tissues. Three miR-124 encoding genes, including mir-124-1, mir-124-2, and mir-124-3, harboring CpG islands undergo methylation-mediated miR-124 inactivation in ESCC tissues. The methylation status of all these three genes was negatively associated with the expression of miR-124. The low expression of miR-124 and the hypermethylation of mir-124-1 and mir-124-3 were associated with the clinico-pathological parameters indicating the poor prognosis. In addition, promoter methylation of all three genes plus low expression of miR-124 was the independent poor prognostic marker for ESCC patients. In conclusion, miR-124 may function as a tumor suppressive miRNA, and hypermethylation-mediated inactivation of miR-124 may be useful for a poor prognostic marker for ESCC patients.






Similar content being viewed by others
Abbreviations
- miRNAs:
-
microRNAs
- ESCC:
-
Esophageal squamous cell carcinoma
- EAC:
-
Esophageal adenocarcinoma
- 3′UTRs:
-
3′ Untranslated regions
References
Siegel RL et al (2016) Cancer statistics, 2016. CA Cancer J Clin 66:7–30
Kaz AM et al (2014) Epigenetic biomarkers in esophageal cancer. Cancer Lett 342:193–199
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
He L et al (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5:522–531
Di Leva G et al (2014) MicroRNAs in cancer. Annu Rev Pathol 9:287–314
Samantarrai D et al (2013) Genomic and epigenomic cross-talks in the regulatory landscape of miRNAs in breast cancer. Mol Cancer Res 11:315–328
Lu L et al (2007) Hypermethylation of let-7a-3 in epithelial ovarian cancer is associated with low insulin-like growth factor-II expression and favorable prognosis. Cancer Res 67:10117–10122
Han L et al (2007) DNA methylation regulates microRNA expression. Cancer Biol Ther 6:1284–1288
Lujambio A et al (2008) A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A 105:13556–13561
Wang Y et al (2016) miR-124-3p functions as a tumor suppressor in breast cancer by targeting CBL. BMC Cancer 16:826
Geng S et al (2014) The tumor suppressor role of miR-124 in osteosarcoma. PLoS ONE 9:e91566
Ueda Y et al (2014) DNA methylation of microRNA-124a is a potential risk marker of colitis-associated cancer in patients with ulcerative colitis. Dig Dis Sci 59:2444–2451
Wang H et al (2017) Pyrosequencing quantified methylation level of miR-124 predicts shorter survival for patients with myelodysplastic syndrome. Clin Epigenet 9:91
Mathé EA et al (2009) MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res 15:6192–6200
Sauvageau M et al (2010) Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell 7:299–313
Dong F et al (2017) Dysregulation of miRNAs in bladder cancer: altered expression with aberrant biogenesis procedure. Oncotarget 8:27547–27568
Jafri MA et al (2017) Role of miRNAs in human cancer metastasis: implications for therapeutic intervention. Semin Cancer Biol 44:117–131
Ma T et al (2016) MicroRNA-124 functions as a tumor suppressor by regulating CDH2 and epithelial-mesenchymal transition in non-small cell lung cancer. Cell Physiol Biochem 38:1563–1574
Li W et al (2017) miR-124 acts as a tumor suppressor in glioblastoma via the inhibition of signal transducer and activator of transcription 3. Mol Neurobiol 54:2555–2561
Zhao Y et al (2017) MiR-124 acts as a tumor suppressor by inhibiting the expression of sphingosine kinase 1 and its downstream signaling in head and neck squamous cell carcinoma. Oncotarget 8:25005–25020
Wang P et al (2014) Methylation-mediated silencing of the miR-124 genes facilitates pancreatic cancer progression and metastasis by targeting Rac1. Oncogene 33:514–524
Wilting SM et al (2010) Methylation-mediated silencing and tumour suppressive function of hsa-miR-124 in cervical cancer. Mol Cancer 9:167
Gebauer K et al (2013) Hsa-mir-124-3 CpG island methylation is associated with advanced tumours and disease recurrence of patients with clear cell renal cell carcinoma. Br J Cancer 108:131–138
Asada K et al (2015) Demonstration of the usefulness of epigenetic cancer risk prediction by a multicentre prospective cohort study. Gut 64:388–396
Liu F et al (2016) Aberrant overexpression of EZH2 and H3K27me3 serves as poor prognostic biomarker for esophageal squamous cell carcinoma patients. Biomarkers 21:80–90
Okamoto K et al (2016) Expression status of CD44 and CD133 as a prognostic marker in esophageal squamous cell carcinoma treated with neoadjuvant chemotherapy followed by radical esophagectomy. Oncol Rep 36:3333–3342
Lin Y et al (2017) P21, COX-2, and E-cadherin are potential prognostic factors for esophageal squamous cell carcinoma. Dis Esophagus 30:1–10
Wang Y et al (2017) MicroRNA expression in esophageal squamous cell carcinoma: novel diagnostic and prognostic biomarkers. Mol Med Rep 15:3833–3839
Wu X et al (2017) NORAD expression is associated with adverse prognosis in esophageal squamous cell carcinoma. Oncol Res Treat 40:370–374
Yang XZ et al (2018) Predictive value of LINC01133 for unfavorable prognosis was impacted by alcohol in esophageal squamous cell carcinoma. Cell Physiol Biochem 48:251–262
Kang K et al (2018) Expression of UCA1 and MALAT1 long-chain non-coding RNAs in esophageal squamous cell carcinoma tissues is predictive of patient prognosis. Arch Med Sci 14:752–759
Bao J et al (2018) Upregulation of the long noncoding RNA FOXD2-AS1 predicts poor prognosis in esophageal squamous cell carcinoma. Cancer Biomark 21:527–533
Xia W et al (2016) Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep 6:35576
Agirre X et al (2009) Epigenetic silencing of the tumor suppressor microRNA HsamiR-124a regulates CDK6 expression and confers a poor prognosis in acute lymphoblastic leukemia. Cancer Res 69:4443–4453
Shi XB et al (2015) miR-124 and androgen receptor signaling inhibitors repress prostate cancer growth by downregulating androgen receptor splice variants, EZH2, and Src. Cancer Res 75:5309–5317
Zheng F et al (2012) The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut 61:278–289
Qiao W et al (2017) miR-124 suppresses glioblastoma growth and potentiates chemosensitivity by inhibiting AURKA. Biochem Biophys Res Commun 486:43–48
Liu S et al (2016) miR-124 inhibits proliferation and invasion of human retinoblastoma cells by targeting STAT3. Oncol Rep 36:2398–2404
Acknowledgements
This work was supported by the Grants from the Ordinary University Considerable Distinctive Subjects Foundation of Hebei Province (No. 200552).
Funding
This work was supported by National Nature Science Foundation of China (No. 81001178), and Science Foundation of Hebei Province (No. 16967788D).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors had no conflicts of interest to declare in relation to this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10585_2019_9974_MOESM1_ESM.tif
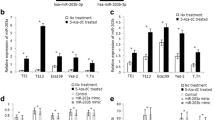
Supplementary Fig. 1. The expression of miR-124-3p and EZH2 in ESCC cell lines. (A) The expression of miR-124-3p and EZH2 in four ESCC cell lines was detected by qRT-PCR. (B) The correlation between EZH2 and miR-124-3p expression in above four ESCC cell lines. Supplementary material 1 (TIFF 24 kb)
10585_2019_9974_MOESM2_ESM.png
Supplementary Fig. 2. The expression of miR-124 using 5S rRNA as an internal reference gene in primary tumor specimens. (A) Relative expression of miR-124-3p in ESCC tissues and corresponding normal tissues. Data are expressed as the Mean ± SEM of three independent replicate experiments (6.32 ± 0.21 vs. 3.12 ± 0.14, P < 0.001). (B) The expression of miR-124-3p was correlated with the tumor infiltration, lymphode metastasis, distant metastasis or recurrence, and clinical stage. Data are expressed as the Mean ± SEM of three independent replicate experiments (4.52 ± 0.20 vs. 2.24 ± 0.11, P < 0.001; 4.42 ± 0.18 vs. 3.26 ± 0.17, P < 0.01; 3.72 ± 0.12 vs. 2.17 ± 0.11, P < 0.001; 3.86 ± 0.16 vs. 2.37 ± 0.14, P < 0.001). (C) Relative expression of miR-124 in mir-124-1, mir-124-2, mir-124-3 methylation or unmethylation ESCC tissues. Data are expressed as the Mean ± SEM of three independent replicate experiments (4.16 ± 0.27 vs. 2.95 ± 0.15, P < 0.001; 4.43 ± 0.27 vs. 3.06 ± 0.13, P < 0.001; 4.97 ± 0.26 vs. 3.13 ± 0.14, P < 0.001). Supplementary material 2 (PNG 58 kb)
Rights and permissions
About this article
Cite this article
Tian, Z., Li, Z., Zhu, Y. et al. Hypermethylation-mediated inactivation of miR-124 predicts poor prognosis and promotes tumor growth at least partially through targeting EZH2/H3K27me3 in ESCC. Clin Exp Metastasis 36, 381–391 (2019). https://doi.org/10.1007/s10585-019-09974-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10585-019-09974-1