Abstract
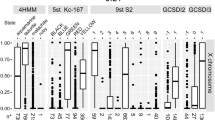
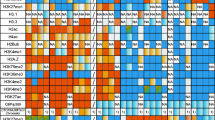
Historically, chromatin has been subdivided into heterochromatin, transcriptionally inactive regions that remain densely packaged throughout the cell cycle, and euchromatin, transcriptionally active regions that take on a diffuse appearance as the cell enters interphase. The banded portion of the small fourth chromosome (dot chromosome) of Drosophila melanogaster is unusual in exhibiting many characteristics of heterochromatic domains, and at the same time maintaining a gene density typical of euchromatin. Similar to genes embedded in pericentric heterochromatin, many of the dot chromosome genes have adapted to a heterochromatic environment. Little is known about the regulation of these genes and less about their evolution in a chromatin context. Interestingly, most of the genes from the D. melanogaster fourth chromosome remain clustered on a small chromosome throughout the genus Drosophila; yet the dot chromosome appears euchromatic in some species, such as D. virilis. Existing genomic sequence data allow an exploration of the underlying differences in DNA sequence organization between species. Here we review the available data describing the dot chromosome, which derives primarily from D. melanogaster. With its unusual and changing nature, the dot chromosome in the genus Drosophila provides a unique opportunity for the examination of transitions between chromatin states during evolution.
Similar content being viewed by others
References
Ashburner M, Golic KG, Hawley RS (2005) Drosophila: A Laboratory Handbook. Cold Spring Harbor: Cold Spring Harbor Press.
Bartolome C, Maside X, Charlesworth B (2002) On the abundance and distribution of transposable elements in the genome of Drosophila melanogaster. Mol Biol Evol 19: 926–937.
Carvalho AB, Clark AG (2005) Y chromosome of D. pseudoobscura is not homologous to the ancestral Drosophila Y. Science 307: 108–110.
Copenhaver GP, Nickel K, Kuromori T et al. (1999) Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286: 2468–2474.
Czermin B, Melfi R, McCabe D et al. (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185–196.
Eissenberg JC, Elgin SC (2000) The HP1 protein family: getting a grip on chromatin. Curr Opin Genet Dev 10: 204–210.
Fanti L, Berloco M, Piacentini L, Pimpinelli S (2003) Chromosomal distribution of heterochromatin protein 1 (HP1) in Drosophila: a cytological map of euchromatic HP1 binding sites. Genetica 117: 135–147.
Franke A, Baker BS (1999) The rox1 and rox2 RNAs are essential components of the compensasome, which mediates dosage compensation in Drosophila. Mol Cell 4: 117–122.
Grewal SI, Elgin SC (2002) Heterochromatin: new possibilities for the inheritance of structure. Curr Opin Genet Dev 12: 178–187.
Haynes KA, Leibovitch BA, Rangwala SH, Craig C, Elgin SC (2004) Analyzing heterochromatin formation using chromosome 4 of Drosophila melanogaster. Cold Spring Harbor Symp Quant Biol 69: 267–272.
Hearn MG, Hedrick A, Grigliatti TA, Wakimoto BT (1991) The effect of modifiers of position-effect variegation on the variegation of heterochromatic genes of Drosophila melanogaster. Genetics 128: 785–797.
Hochman B (1976) The fourth chromosome of Drosophila melanogaster. In: Ashburner M, Novitski E, eds. The Genetics and Biology of Drosophila. New York: Academic Press, pp. 903–928.
Hoskins RA, Smith CD, Carlson JW et al. (2002) Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol 3: Research0085.
Jensen MA, Charlesworth B, Kreitman M (2002) Patterns of genetic variation at a chromosome 4 locus of Drosophila melanogaster and D. simulans. Genetics 160: 493–507.
Kikkawa H (1938) Studies on the genetics and cytology of Drosophila ananassae. Genetica 20: 458–516.
Krivshenko J (1952) A cytogenetic study of the Y chromosome in Drosophila busckii. Genetics 37: 500–518.
Krivshenko J (1955) A cytogenetic study of the X chromosome of Drosophila busckii and its relation to phylogeny. Genetics 41: 1071–1079.
Krivshenko J (1959) New evidence for the homology of the short euchromatic elements of the X and Y chromosomes of Drosophila busckii with the microchromosome of Drosophila melanogaster. Genetics 44: 1027–1040.
Larsson J, Chen JD, Rasheva V, Rasmuson-Lestander A, Pirrotta V (2001) Painting of fourth, a chromosome-specific protein in Drosophila. Proc Natl Acad Sci USA 98: 6273–6278.
Larsson J, Svensson MJ, Stenberg P, Makitalo M (2004) Painting of fourth in genus Drosophila suggests autosome-specific gene regulation. Proc Natl Acad Sci USA 101: 9728–9733.
Lindsley DL, Sandler L, Baker BS et al. (1972) Segmental aneuploidy and the gross structure of the Drosophila genome. Genetics 71: 157–184.
Locke J, McDermid HE (1993) Analysis of Drosophila chromosome 4 using pulsed field gel electrophoresis. Chromosoma 102: 718–723.
Locke J, Podemski L, Roy K, Pilgrim D, Hodgetts R (1999) Analysis of two cosmid clones from chromosome 4 of Drosophila melanogaster reveals two new genes amid an unusual arrangement of repeated sequences. Genome Res 9: 137–149.
Mohr OL (1932) Genetical and cytological proof of somatic elimination of the fourth chromosome in Drosophila melanogaster. Genetics 17: 60–80.
Müller HJ (1940) Bearings of the Drosophila work on systematics. In: Huxley J, eds. The New Systematics. Oxford: Clarendon Press, pp. 185–268.
Nagaki K, Cheng Z, Ouyang S et al. (2004) Sequencing of a rice centromere uncovers active genes. Nat Genet 36: 138–145.
Podemski L, Ferrer C, Locke J (2001) Whole arm inversions of chromosome 4 in Drosophila species. Chromosoma 110: 305–312.
Sandler L, Novitski E (1955) Evidence for genetic homology between chromosomes I and IV in Drosophila melanogaster with a proposed explanation for the crowding effect in triploids. Genetics 41: 189–193.
Sandler L, Szauter P (1978) The effect of recombination-defective meiotic mutants on fourth-chromosome crossing over in Drosophila melanogaster. Genetics 90: 699–712.
Schotta G, Ebert A, Krauss V et al. (2002) Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J 21: 1121–1131.
Schulze SR, Sinclair DA, Fitzpatrick KA, Honda BM (2005) A genetic and molecular characterization of two proximal heterochromatic genes on chromosome 3 of Drosophila melanogaster. Genetics 169: 2165–2177.
Sheldahl LA, Weinreich DM, Rand DM (2003) Recombination, dominance and selection on amino acid polymorphism in the Drosophila genome: contrasting patterns on the X and fourth chromosomes. Genetics 165: 1195–1208.
Slawson EE, Shaffer CD, Malone CD et al. (2006) Comparison of dot chromosome sequences from D. melanogaster and D. virilis reveals an enrichment of DNA transposon sequences in heterochromatic domains. Genome Biol (In press).
Stenberg P, Pettersson F, Saura AO, Berglund A, Larsson J (2005) Sequence signature analysis of chromosome identity in three Drosophila species. BMC Bioinformatics 6: 158.
Sturtevant AH (1934) Preferential segregation of the fourth chromosomes in Drosophila melanogaster. Genetics 20: 515–518.
Sturtevant AH (1936) Preferential segregation in triplo-IV females in Drosophila melanogaster. Genetics 21: 444–466.
Sturtevant AH (1946) On the dot chromosome of Drosophila repleta and D. hydei. Genetics 31: 259–268.
Sun FL, Cuaycong MH, Craig CA et al. (2000) The fourth chromosome of Drosophila melanogaster: interspersed euchromatic and heterochromatic domains. Proc Natl Acad Sci USA 97: 5340–5345.
Sun FL, Cuaycong MH, Elgin SC (2001) Long-range nucleosome ordering is associated with gene silencing in Drosophila melanogaster pericentric heterochromatin. Mol Cell Biol 21: 2867–2879.
Sun FL, Haynes K, Simpson CL et al. (2004) cis-Acting determinants of heterochromatin formation on Drosophila melanogaster chromosome four. Mol Cell Biol 24: 8210–8220.
Wakimoto BT, Hearn MG (1990) The effects of chromosome rearrangements on the expression of heterochromatic genes in chromosome 2L of Drosophila melanogaster. Genetics 125: 141–154.
Wallrath LL, Elgin SC (1995) Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev 9: 1263–1277.
Wang W, Thornton K, Berry A, Long M (2002) Nucleotide variation along the Drosophila melanogaster fourth chromosome. Science 295: 134–137.
Wang W, Thornton K, Emerson JJ, Long M (2004) Nucleotide variation and recombination along the fourth chromosome in Drosophila simulans. Genetics 166: 1783–1794.
Yan H, Jin W, Nagaki K et al. (2005) Transcription and histone modifications in the recombination-free region spanning a rice centromere. Plant Cell 17: 3227–3238.
Yang HP, Hung TL, You TL, Yang TH (2005) Genome-wide comparative analysis of the highly abundant transposable element DINE-1 suggests a recent transpositional burst in Drosophila yakuba. Genetics
Yasuhara JC, DeCrease CH, Wakimoto BT (2005) Evolution of heterochromatic genes of Drosophila. Proc Natl Acad Sci USA 102: 10958–10963.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Riddle, N.C., Elgin, S.C.R. The dot chromosome of Drosophila: Insights into chromatin states and their change over evolutionary time. Chromosome Res 14, 405–416 (2006). https://doi.org/10.1007/s10577-006-1061-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-006-1061-6