Abstract
Objectives
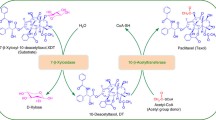
Taxoid 10β-O-acetyl transferase (DBAT) was redesigned to enhance its catalytic activity and substrate preference for baccatin III and taxol biosynthesis.
Results
Residues H162, D166 and R363 were determined as potential sites within the catalytic pocket of DBAT for molecular docking and site-directed mutagenesis to modify the activity of DBAT. Enzymatic activity assays revealed that the kcat/KM values of mutant H162A/R363H, D166H, R363H, D166H/R363H acting on 10-deacetylbaccatin III were about 3, 15, 26 and 60 times higher than that of the wild type of DBAT, respectively. Substrate preference assays indicated that these mutants (H162A/R363H, D166H, R363H, D166H/R363H) could transfer acetyl group from unnatural acetyl donor (e.g. vinyl acetate, sec-butyl acetate, isobutyl acetate, amyl acetate and isoamyl acetate) to 10-deacetylbaccatin III.
Conclusion
Taxoid 10β-O-acetyl transferase mutants with redesigned active sites displayed increased catalytic activities and modified substrate preferences, indicating their possible application in the enzymatic synthesis of baccatin III and taxol.




Similar content being viewed by others
References
Bontpart T, Cheynier V, Ageorges A, Terrier N (2015) BAHD or SCPL acyltransferase? What a dilemma for acylation in the world of plant phenolic compounds. New Phytol 208(3):695–707
Croteau R, Ketchum RE, Long RM, Kaspera R, Wildung MR (2006) Taxol biosynthesis and molecular genetics. Phytochem Rev 5(1):75–97
Currin A, Swainston N, Day PJ, Kell DB (2015) Synthetic biology for the directed evolution of protein biocatalysts: navigating sequence space intelligently. Chem Soc Rev 44(5):1172–1239
Damborsky J, Brezovsky J (2014) Computational tools for designing and engineering enzymes. Curr Opin Chem Biol 19:8–16
Deng W, Wang Y, Liu Z, Cheng H, Xue Y (2014) HemI: a toolkit for illustrating heatmaps. PLoS ONE 9(11):e111988
Egesborg P, Carlettini H, Volpato JP, Doucet N (2015) Combinatorial active-site variants confer sustained clavulanate resistance in BlaC beta-lactamase from Mycobacterium tuberculosis. Protein Sci 24(4):534–544
Han F, Kang LZ, Zeng XL, Ye ZW, Guo LQ, Lin JF (2014) Bioproduction of baccatin III, an advanced precursor of paclitaxol, with transgenic Flammulina velutipes expressing the 10-deacetylbaccatin III-10-O-acetyl transferase gene. J Sci Food Agric 94(12):2376–2383
Li G, Zhang H, Sun Z, Liu X, Reetz MT (2016) Multiparameter optimization in directed evolution: engineering thermostability, enantioselectivity, and activity of an epoxide hydrolase. ACS Catal 6(6):3679–3687
Li BJ, Wang H, Gong T, Chen JJ, Chen TJ, Yang JL, Zhu P (2017) Improving 10-deacetylbaccatin III-10-beta-O-acetyltransferase catalytic fitness for Taxol production. Nat Commun 8:15544
Liu H, Chen Q (2016) Computational protein design for given backbone: recent progresses in general method-related aspects. Curr Opin Struct Biol 39:89–95
Navarro-Retamal C, Gaete-Eastman C, Herrera R, Caballero J, Alzate-Morales JH (2016) Structural and affinity determinants in the interaction between alcohol acyltransferase from F. x ananassa and several alcohol substrates: a computational study. PLoS ONE 11(4):e0153057
Patel RN, Banerjee A, Nanduri VV (2000) Enzymatic acetylation of 10-deacetylbaccatin III to baccatin III by C-10 deacetylase from Nocardioides luteus SC 13913. Enzyme Microb Technol 27(6):371–375
Sandhya S, Mudgal R, Kumar G, Sowdhamini R, Srinivasan N (2016) Protein sequence design and its applications. Curr Opin Struct Biol 37:71–80
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461
Webb B, Sali A (2014) Comparative protein structure modeling using MODELLER. Curr Protoc Bioinform 47:5.6.1–32
Wijma HJ, Floor RJ, Bjelic S, Marrink SJ, Baker D, Janssen DB (2015) Enantioselective enzymes by computational design and in silico screening. Angew Chem Int Ed Engl 54(12):3726–3730
Yamamoto K, Miyake H, Kusunoki M, Osaki S (2011) Steric hindrance by 2 amino acid residues determines the substrate specificity of isomaltase from Saccharomyces cerevisiae. J Biosci Bioeng 112(6):545–550
Zheng L, Baumann U, Reymond JL (2004) An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res 32(14):e115
Acknowledgements
This work was supported by the Science and Technology Program of Guangdong Province (Grant 2014B050505018, 2014B020205003, 2015A020209121) and the National Natural Science Foundation of China (Grant 31071837, 31572178, 31772373).
Supporting information
Supplementary Table 1—Primers used in the experiments.
Supplementary Table 2—DBAT homology model.
Supplementary Fig. 1—Expression of mutants.
Supplementary Fig. 2—The kinetic curves for DBAT.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
You, LF., Huang, JJ., Wei, T. et al. Enhanced catalytic activities and modified substrate preferences for taxoid 10β-O-acetyl transferase mutants by engineering catalytic histidine residues. Biotechnol Lett 40, 1245–1251 (2018). https://doi.org/10.1007/s10529-018-2573-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-018-2573-9