Abstract
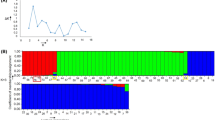
Simple sequence repeat (SSR) markers are the major molecular tools for genetic and genomic researches that have been extensively developed and used in major crops. However, few are available for lentils (Lens culinaris M.), economically an important cool-season legume. The lack of informative simple sequence repeat (SSR) markers in lentil has been a major limitation for lentil molecular breeding studies. Therefore, in order to develop SSR markers for lentil, an enriched genomic libraries for AC and AG repeats were constructed from the Lens culinaris cv Kafkas. A total of 350 clones were inquired for the detection of SSRs. Of 350 clones, 68 had SSR motifs. In polymorphism analysis using 53 newly developed SSRs, a total of 144 alleles across 24 lentil cultivars were detected with an average of 4.64 per locus. The average heterozygosity was 0.588 and polymorphism information contents ranged from 0.194 to 0.895 with an average value of 0.520. These newly developed SSRs will constitute useful tools for molecular breeding, mapping, and assessments of genetic diversity and population structure of lentils.

Similar content being viewed by others
References
Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9:208–218
Andeden EE, Baloch FS, Çakır E, Toklu F, Özkan H (2015) Development, characterization and mapping of microsatellite markers for lentil (Lens culinarisMedik.). Plant Breed 134:589–598
Alghamdi SS, Khan AM, Ammar MH, El-Harty EH, Migdadi HM, Abd El-Khalik SM, Al-Shameri AM, Javed MM, Al-Faifi SA (2014) Phenological, nutritional and molecular diversity assessment among 35 introduced lentil (Lens culinaris Medik.) genotypes grown in Saudi Arabia. Int J Mol Sci 15:277–295
Ates D, Aldemir S, Alsaleh A, Erdogmus S, Nemli S, Kahriman A, Ozkan H, Vandenberg A, Tanyolac B (2018) A consensus linkage map of lentil based on DArT markers from three RIL mapping populations. PLoS ONE 13(1):e0191375. https://doi.org/10.1371/journal.pone.0191375
Bloor PA, Barker FS, Watts PC, Noyes HA, Kemp SJ (2001) Microsatellite libraries by enrichment. https://www.genomics.liv.ac.uk/animal/RESEARCH/MICROSAT.PDF Accessed 10 April 2018
Cubero JI (1981) Taxonomy, distribution and evolution of the lentil and its wild relatives. In: Witcombe J, Erskine W (eds) Assessment of genetic diversity in lentils (Lens culinaris Medik) based on SNPs. M. Nijhoff & W. Junk Publisher, Boston, pp 187–204
Cuc LM, Mace ES, Crouch JH, Quang VD, Long TD, Varshney RK (2008) Isolation and characterization of novel microsatellite markers and their application for diversity assessment in cultivated groundnut (Arachis hypogaea). BMC Plant Biol 8:55
Dikshit HK, Singh A, Singh D, Aski MS, Prakash P, Jain N, Meena S, Kumar S, Sarker A (2015) Genetic diversity in Lens species revealed by EST and genomic simple sequence repeat analysis. PLoS ONE 10:0138101. https://doi.org/10.1371/journal.pone.0138101
El-Nahas A, El-Shazly H, Ahmed S, Omran A (2011) Molecular and biochemical markers in some lentil (Lens culinaris Medik.) genotypes. Ann Agric Sci 56:105–112
Eujayl IM, Baum M, Powell W, Erskine W, Pehu E (1998) Agenetic linkage map of lentil (Lens sp.) based on RAPD and AFLP markers using recombinant inbred lines. Theor Appl Genet 97:83–89
FAOSTAT (2013) https://faostat3.fao.org/faostat-gateway/go/to/download/Q/QC/E. Accessed 7 April 2018
Ferguson ME, Robertson LD, Ford-Lloyd BV, Newbury HJ, Maxted N (1998) Contrasting genetic variation amongst lentil landraces from different geographical origins. Euphytica 102:265–273
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Havey MJ, Muehlbauer FJ (1989) Variability for restriction fragment lengths and phylogenies in lentil. Theor Appl Genet 77:839–843
Hendre PS, Aggarwal RK (2007) DNA Markers: development and application for genetic improvement of coffee. In: Varshney RK, Tuberosa R (eds) Genomics-assisted crop improvement. Springer, Dordrecht, pp 399–434
Huang X, Madan A (1999) CAP3: A DNA sequence assembly program. Genome Resources 9:868–877
Hamwieh A, Udupa SM, Sarkar A, Jung C, Baum M (2009) Development of new microsatellite markers and their application in the analysis of genetic diversity in lentils. Breed Sci 59:77–86
Hamwieh A, Udupa SM, Choumane W, Sarker A, Dreyer F, Jung C, Baum M (2005) A genetic linkage map of Lens sp. based on microsatellite and AFLP markers and the localization of fusarium vascular wilt resistance. Theor Appl Genet 110:669–677
Idrissi O, Udupa SM, Houasli C, De Keyser E, Van Damme P, De Riek J (2015) Genetic diversity analysis of Moroccan lentil (Lens culinaris Medik.) landraces using simple sequence repeat and amplified fragment length polymorphisms reveals functional adaptation towards agro-environmental origins. Plant Breed 134:322–332
Koressaar T, Remm M (2007) Enhancements and modifications of primer design program Primer3. Bioinformatics 2310:1289–1291
Kahraman A, Kusmenoglu I, Ayidn N, Aydogan A, Erskin W, Muehlbauer FJ (2004) QTL mapping of winter hardiness genes in lentil. Crop Sci 44:13–22
Kumar S, Hamweih A, Manickavelu A, Kumar J, Sharma TR, Baum M (2014) Advances in lentil genomics. In: Gupta S, Nadarajan N, Gupta DS (eds) Legumes in Omics Era. Springer Science+Business Media, New York, pp 111–130
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Lefort F, Lally M, Thompson D, Douglas GC (1998) Morphological traits, microsatellite fingerprinting and genetic relatedness of a stand of elite oaks (Q. robur L.) at Tullynally Ireland. Silvae Genet 47:5–6
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci 70:3321–3323
Powell W, Mackray GC, Provan J (1996) Polymorphism revealed by simple sequence repeats. Trends in Plant Sci 1:215–222
Phan HTT, Ellwood SR, Hane JK, Ford R, Materne M, Oliver RP (2007) Extensive macrosynteny between Medicago truncatula and Lens culinaris ssp. culinaris. Theo Appl Genet 114:549–558
Schuelke M (2000) An economic method for the fluorescent labelling of PCR fragments. Nat Biotechnol 18:233–234
Sonnante G, Pignone D (2001) Assessment of genetic variation in a collection of lentil using molecular tools. Euphytica 120:301–307
Sharma SK, Knox MR, Ellis THN (1996) AFLP analysis of the diversity and phylogeny of Lens and its comparison with RAPD analysis. Theor Appl Genet 93:751–758
Saha GC, Sarker A, Chen W, Vandemark GJ, Muehlbauer FJ (2013) Inheritance and linkage map positions of genesconferring agromorphological traits in Lens culinaris Medik. Int J Agron 618926 9:pages. https://doi.org/10.1155/2013/618926
Tsanakas GF, Mylona PV, Koura K, Polidoros AN, Gleridou A (2018) Genetic diversity analysis of the Greek lentil (Lens culinaris) landrace ‘Eglouvis’ using morphological and molecular markers. Plant Genet Resour. https://doi.org/10.1017/S147926211800009610.1017/S1479262118000096
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tanyolac B, Ozatay S, Kahraman A, Muehlbauer F (2010) Linkage mapping of lentil (Lens culinaris L.) genome using recombinant inbred lines revealed by AFLP, ISSR, RAPD and some morphologic markers. J Agric Biotechnol Sustain Dev 2:001–006
Techen N, Arias RS, Glynn NC, Pan Z, Khan IA, Scheffler BE (2010) Optimized construction of microsatellite-enriched libraries. Mol Ecol Resour 10:508–515
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch SR (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 11:1441–1452
Toklu F, Karaköy T, Hakle I, Bicer T, Brandolini A, Kilian B, Ozkan H (2009) Genetic variation among lentil (Lens culinaris Medik.) landraces from Southeast Turkey. Plant Breed. 128:178–186
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40(15):e115
Verma P, Sharma TR, Srivastava PS, Abdin MZ, Bhatia S (2014) Exploring genetic variability within lentil (Lens culinaris Medik.) and across related legumes using a newly developed set of microsatellite markers. Mol Biol Rep 41:5607–5625
You FM, Huo N, Gu YQ, Luo MC, Ma Y, Hane D, Lazo GR, Dvorak J, Anderson OD (2008) BatchPrimer3: a high throughput web application for PCR and sequencing primer designi. BMC Bioinform 9:253. https://doi.org/10.1186/1471-2105-9-253
Acknowledgements
This study was supported by TUBITAK (The Scientific and Technical Research Council of Turkey, TOVAG-215O088) and Erciyes University, Scientific Research Projects Unit (FDA-2017–7099). Thanks are extended to Betül-Ziya Eren Genome and Stem Cell Center of Erciyes University for their supports provided throughout the experiments. The authors are also grateful to Assoc. Prof. Dr.Zeki Gökalp (a Certified English Translator and an expert in Biosystems Engineering) for his critical reading and through syntactic corrections of the manuscript.
Funding
This study was funded by TUBITAK (Grant Number 215O088) and Erciyes University Scientific Research Projects Coordination Unit (Grant Number FDA-2017–7099).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Bakır, M., Kahraman, A. Development of New SSR (Simple Sequence Repeat) Markers for Lentils (Lens culinaris Medik.) from Genomic Library Enriched with AG and AC Microsatellites. Biochem Genet 57, 338–353 (2019). https://doi.org/10.1007/s10528-018-9893-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-018-9893-2