Abstract
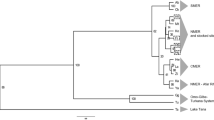
Red tilapia is thought to be the result of mutant-colored female Mozambique tilapias mating with male Nile, blue, or Zanzibar tilapias and cultured widely in Asia and central and South America. However, there is limited information about its genetic diversity and stock structure. In this study, we investigated the genetic variability of red tilapia stocks to provide fundamental knowledge for genetic improvement by molecular-marker-assisted selective breeding programs. Individuals (n = 180) from three stocks (Chinese Taiwan, Israel, and Malaysia) of red tilapia were genotyped based on 14 microsatellite markers. The results showed that all microsatellite loci were detected with high levels of polymorphism, with a mean number of 14.87 ± 3.85 alleles per locus in all stocks. Taiwan and Israel stocks showed higher heterozygosity than did the Malaysia stock. The F-statistic analysis showed that there was no significant genetic differentiation between the Taiwan and Israel stocks (P > 0.05), whereas there was highly significant genetic divergence in the other pairwise stocks (P < 0.01), suggesting that Taiwan and Israel stocks could be regarded as a single genetic group distinct from Malaysia stock. This result was in accordance with the UPGMA dendrogram based on Nei’s genetic distances of three stocks. The analysis of molecular variances (AMOVAs) revealed highly significant genetic variation among three stocks (P < 0.01) and accounted for 8.68% of the total variance. The results reported above were confirmed by Bayesian analysis in genetic structure simulation, which indicated a distinct genetic difference between Taiwan and Israel stocks compared with Malaysia stock.



Similar content being viewed by others
References
Balloux F, Lugon-Moulin N (2002) The estimation of population differentiation with microsatellite markers. Mol Ecol 11(22):155–165
Botsein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Chen YS, Dai CJ (2003) Evaluation onnutritional components of the muscle in the Oreochromis niloticus×O.mossambicus. Journal of Fujian Normal University (natural science) 19(4): 62-66Cornuet JM, Luikart G (1996) description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1:47–50
FAO (2013) Food and Agriculture Organization of the United Nations (FAO). Fisheries and aquaculture department, Global Aquaculture Production 1950–2013
Frankham R (1996) Relationship of genetic variation to population size in wildlife. Conserv Biol 10:1500–1508
Galman OR, Avtalion RR (1983) A preliminary investigation of the characteristics of red tilapias from the Philippines and Taiwan. In: Fishelson, L, Yaron, Z (comps), proceedings of the international symposium on tilapia in aquaculture 8-13
Ha HP, Nguyen TTT, Poompuang S, Na-Nakorn U (2009) Microsatellites revealed no genetic differentiation between hatchery and contemporary wildpopulations of striped catfish, Pangasianodon hypophthalmus (Sauvage 1878) in Vietnam. Aquaculture 291:154–160
Kaczmarczyk D, Luczynski M, Brzuzan P (2012) Genetic variation in three paddlefish (Polyodon spathula Walbaum) stocks based on microsatellite DNA analysis. Czech J Anim Sci 57(8):345–352
Kalinowski ST (2002) How many alleles per locus should be used to estimate genetic distance? Heredity 88(1):62–65
Kalinowski ST (2005) Dopolymorphic loci require large sample sizes to estimate genetic distance? Heredity 94(1):33–36
Kalinowski ST, Taper ML, Marshall TC (2007) Revising how the computer program cervus accommodates genotyping error increases success in paternity assignment. Mol Ecol 16:1099–1106
Kocher TD, Lee WJ, Sobolewaka H, Penman D, Mcandrew B (1998) A genetic linkage map of a cichlid fish, the tilapia (Oreochromis niloticus). Genetics 148:1225–1232
Li JL, Tang YK, Chen WH, Yu JH, Dong ZJ, Xu P (2009) Association analysis of microsatellite DNA markers with body weight and body shape in GIFT. J Fish Sci China 16(6):824–832
Liu ZJ, Cordes JF (2004) DNA marker technologies and their applications in aquaculture genetics. Aquaculture 238(1–4):1–37
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89(3):583–590
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York, pp 159–164
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multi-locus data. Genetics 155:945–959
Romana-Eguia MRR, Eguia RV (1999) Growth of five Asian red tilapia strains in saline environments. Aquaculture 173:161–170
Romana-Egui MRR, Ikeda M, Basiao ZU, Taniguchi N (2004) Genetic diversity in farmed Asian Nile and red hybrid tilapia stocks evaluated from microsatellite and mitochondrial DNA analysis. Aquaculture 236:131–150
Rousset F (2008) GENEPOP’007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour 8:103–106
Sandeep M, Sun F, Liu F, Li J, David PB, Yue GH (2012) Novel polymorphic microsatellites from Florida red tilapia and cross-species amplification in Mozambique and Nile tilapia. J Genet 91:e97–e99
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetic analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Thorp JP (1982) The molecular dock hypothesis: biochemical evolution, genetic differentiation, and systematic. Annu Rev Ecol Syst 13:139–168
Trewavas E (1983) Tilapiine fishes of the genera Sarotherodon, Oreochromis and Danakilia. British Museum (Natural History), London, England
Wohlfarth GW, Rothbard S, Hulata G, Szweigman D (1990) Inheritance of red body coloration in Taiwanese tilapias and in Oreochromis mossambicus. Aquaculture 84:219–234
Yeh FC, Yang RC, Boyle T, Ye ZH, Mao JX (1997) POPGENE, the user-friendly shareware for population genetic analysis. Molecular biology and biotechnology centre. University of Alberta, Canada
Yang H, Zhu WB, Dong ZJ, L F, Gong CP, Liu N, Yuan XH (2015) Morphological variation analysis of three populations of red tilapia. J Shanghai Ocean Univ 24(5):678–684
Zhao MZ, Li Q, Qin Q, Cai YX, Xu ZQ, Wang MH, Chen XH, Ge JC, Bian WJ (2011) Genetic diversity based on microsatellite markers in five channel catfish (Ictalurus punctatus) populations. J Aquac 32(1):24–30
Zhang T, Lu MX, Ye X, Quan YC, Gao FY, Huang ZH, Bai JJ (2009) The genetic structure of four populations of Oreochromis aureus by microsatellite DNA analysis. Acta Hydrobiol Sin (in Chinese) 33:498–508
Zhang YD, Gan X, Tang ZS, Chen Z, Su XH, Tang ZY, Li LP, Lin Y (2010) Analysis of genetic diversity in six tilapia populations. J Northwest A & F Univ (Nat Sci Ed) 38(10):58–66
Acknowledgements
This work was supported by the National Fund for Basic R&D of Non-Profit Research Institutes of China [grant number 2015JBFM07].
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Zhu, W., Yang, H., Yuan, X. et al. High genetic diversity and differentiation in three red tilapia stocks revealed by microsatellite DNA marker analysis. Aquacult Int 25, 1997–2006 (2017). https://doi.org/10.1007/s10499-017-0150-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10499-017-0150-1