Abstract
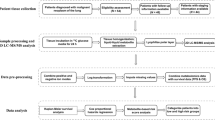
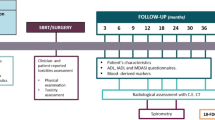
The standard of care for intermediate (Stage II) and advanced (Stages III and IV) non-small cell lung cancer (NSCLC) involves chemotherapy with taxane/platinum derivatives, with or without radiation. Ideally, patients would be screened a priori to allow non-responders to be initially treated with second-line therapies. This evaluation is non-trivial, however, since tumors behave as complex multiscale systems. To address this need, this study employs a multiscale modeling approach to evaluate first-line chemotherapy response of individual patient tumors based on metabolomic analysis of tumor core biopsies obtained during routine clinical evaluation. Model parameters were calculated for a patient cohort as a function of these metabolomic profiles, previously obtained from high-resolution 2DLC-MS/MS analysis. Evaluation metrics were defined to classify patients as Disease-Control (DC) [encompassing complete-response (CR), partial-response (PR), and stable-disease (SD)] and Progressive-Disease (PD) following first-line chemotherapy. Response was simulated for each patient and compared to actual response. The results show that patient classifications were significantly separated from each other, and also when grouped as DC vs. PD and as CR/PR vs. SD/PD, by fraction of initial tumor radius metric at 6 days post simulated bolus drug injection. This study shows that patient first-line chemotherapy response can in principle be evaluated from multiscale modeling integrated with tumor tissue metabolomic data, offering a first step towards individualized lung cancer treatment prognosis.





Similar content being viewed by others
References
Bamji-Stocke, S., V. van Berkel, D. M. Miller, and H. B. Frieboes. A review of metabolism-associated biomarkers in lung cancer diagnosis and treatment. Metabolomics. 14(6):81, 2018.
Bearer, E. L., J. S. Lowengrub, H. B. Frieboes, Y. L. Chuang, F. Jin, S. M. Wise, et al. Multiparameter computational modeling of tumor invasion. Cancer Res. 69(10):4493–4501, 2009.
Buettner, R., J. Wolf, and R. K. Thomas. Lessons learned from lung cancer genomics: the emerging concept of individualized diagnostics and treatment. J. Clin. Oncol. 31(15):1858–1865, 2013.
Cardarella, S., T. M. Ortiz, V. A. Joshi, M. Butaney, D. M. Jackman, D. J. Kwiatkowski, et al. The introduction of systematic genomic testing for patients with non-small-cell lung cancer. J. Thorac. Oncol. 7(12):1767–1774, 2012.
Chen, Y., Z. Ma, A. Li, H. Li, B. Wang, J. Zhong, et al. Metabolomic profiling of human serum in lung cancer patients using liquid chromatography/hybrid quadrupole time-of-flight mass spectrometry and gas chromatography/mass spectrometry. J. Cancer Res. Clin. Oncol. 141(4):705–718, 2015.
Cristini, V., H. B. Frieboes, R. Gatenby, S. Caserta, M. Ferrari, and J. Sinek. Morphologic instability and cancer invasion. Clin. Cancer Res. 11(19 Pt 1):6772–6779, 2005.
Curtis, L. T., C. G. England, M. Wu, J. Lowengrub, and H. B. Frieboes. An interdisciplinary computational/experimental approach to evaluate drug-loaded gold nanoparticle tumor cytotoxicity. Nanomedicine (Lond). 11(3):197–216, 2016.
Curtis, L. T., V. H. van Berkel, and H. B. Frieboes. Pharmacokinetic/pharmacodynamic modeling of combination-chemotherapy for lung cancer. J. Theor. Biol. 448:38–52, 2018.
Daghir-Wojtkowiak, E., P. Wiczling, M. Waszczuk-Jankowska, R. Kaliszan, and M. J. Markuszewski. Multilevel pharmacokinetics-driven modeling of metabolomics data. Metabolomics. 13(3):31, 2017.
Fan, T. W., A. N. Lane, R. M. Higashi, M. Bousamra 2nd., G. Kloecker, and D. M. Miller. Metabolic profiling identifies lung tumor responsiveness to erlotinib. Exp. Mol. Pathol. 87(1):83–86, 2009.
Frieboes, H. B., X. Zheng, C. H. Sun, B. Tromberg, R. Gatenby, and V. Cristini. An integrated computational/experimental model of tumor invasion. Cancer Res. 66(3):1597–1604, 2006.
Frieboes, H. B., L. T. Curtis, M. Wu, K. Kani, and P. Mallick. Simulation of the protein-shedding kinetics of a fully vascularized tumor. Cancer Inform. 14:163–175, 2015.
Govindan, R., L. Ding, M. Griffith, J. Subramanian, N. D. Dees, K. L. Kanchi, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 150(6):1121–1134, 2012.
Hori, S., S. Nishiumi, K. Kobayashi, M. Shinohara, Y. Hatakeyama, Y. Kotani, et al. A metabolomic approach to lung cancer. Lung Cancer. 74(2):284–292, 2011.
Leighl, N. B. Treatment paradigms for patients with metastatic non-small-cell lung cancer: first-, second-, and third-line. Curr. Oncol. 19(Suppl 1):S52–S58, 2012.
Leonard, F., L. T. Curtis, P. Yesantharao, T. Tanei, J. F. Alexander, M. Wu, et al. Enhanced performance of macrophage-encapsulated nanoparticle albumin-bound-paclitaxel in hypo-perfused cancer lesions. Nanoscale. 8(25):12544–12552, 2016.
Leonard, F., L. T. Curtis, M. J. Ware, T. Nosrat, X. Liu, K. Yokoi, et al. Macrophage polarization contributes to the anti-tumoral efficacy of mesoporous nanovectors loaded with albumin-bound paclitaxel. Front. Immunol. 8:693, 2017.
Leonard, F., L. T. Curtis, A. R. Hamed, C. Zhang, E. Chau, D. Sieving, et al. Nonlinear response to cancer nanotherapy due to macrophage interactions revealed by mathematical modeling and evaluated in a murine model via CRISPR-modulated macrophage polarization. Cancer Immunol. Immunother. 69:731, 2020.
Lovly, C., Horn, L., Pao, W. Molecular Profiling of Lung Cancer. My Cancer Genome 2018 [cited 2018 October 1, 2018]. Available from: https://www.mycancergenome.org/content/disease/lung-cancer/
Macklin, P., S. McDougall, A. R. Anderson, M. A. Chaplain, V. Cristini, and J. Lowengrub. Multiscale modelling and nonlinear simulation of vascular tumour growth. J. Math. Biol. 58(4–5):765–798, 2009.
Mahlbacher, G., L. T. Curtis, J. Lowengrub, and H. B. Frieboes. Mathematical modeling of tumor-associated macrophage interactions with the cancer microenvironment. J. Immunother. Cancer. 6:10, 2018.
Marx, V. Biology: the big challenges of big data. Nature. 498(7453):255–260, 2013.
Masters, G. A., S. Temin, C. G. Azzoli, G. Giaccone, S. Baker Jr., J. R. Brahmer, et al. Systemic therapy for stage IV non-small-cell lung cancer: american society of clinical oncology clinical practice guideline update. J. Clin. Oncol. 33(30):3488–3515, 2015.
McDougall, S. R., A. R. Anderson, and M. A. Chaplain. Mathematical modelling of dynamic adaptive tumour-induced angiogenesis: clinical implications and therapeutic targeting strategies. J. Theor. Biol. 241(3):564–589, 2006.
Medina, M. A. Mathematical modeling of cancer metabolism. Crit. Rev. Oncol. Hematol. 124:37–40, 2018.
Mendoza-Juez, B., A. Martinez-Gonzalez, G. F. Calvo, and V. M. Perez-Garcia. A mathematical model for the glucose-lactate metabolism of in vitro cancer cells. Bull. Math. Biol. 74(5):1125–1142, 2012.
Miller, H. A., and H. B. Frieboes. Pharmacokinetic/pharmacodynamics modeling of drug-loaded PLGA nanoparticles targeting heterogeneously vascularized tumor tissue. Pharm. Res. 36(12):185, 2019.
Miller, H. A., and H. B. Frieboes. Evaluation of drug-loaded gold nanoparticle cytotoxicity as a function of tumor vasculature-induced tissue heterogeneity. Ann. Biomed. Eng. 47(1):257–271, 2019.
Miller, H. A., R. Emam, C. M. Lynch, S. Bockhorst, and H. B. Frieboes. Discrepancies in metabolomic biomarker identification from patient-derived lung cancer revealed by combined variation in data pre-treatment and imputation methods. Metabolomics. 17(4):37, 2021.
Miller, H. A., X. Yin, S. A. Smith, X. Hu, X. Zhang, J. Yan, et al. Evaluation of disease staging and chemotherapeutic response in non-small cell lung cancer from patient tumor-derived metabolomic data. Lung Cancer. 156:20–30, 2021.
Miller, H. A., V. van Berkel, and H. B. Frieboes. Lung cancer survival prediction and biomarker identification with an ensemble machine-learning analysis of tumor core biopsy metabolomic data. Metabolomics. 18(8):57, 2022.
Miller, H. A., J. Lowengrub, and H. B. Frieboes. Modeling of tumor growth with input from patient-specific metabolomic data. Ann. Biomed. Eng. 50(3):314–329, 2022.
Miller, H. A., S. N. Rai, X. Yin, X. Zhang, J. A. Chesney, V. van Berkel, et al. Lung cancer metabolomic data from tumor core biopsies enables risk-score calculation for progression-free and overall survival. Metabolomics. 18(5):31, 2022.
Peng, F., Y. Liu, C. He, Y. Kong, Q. Ouyang, X. Xie, et al. Prediction of platinum-based chemotherapy efficacy in lung cancer based on LC-MS metabolomics approach. J. Pharm. Biomed. Anal. 154:95–101, 2018.
Roy, M., and S. D. Finley. Computational model predicts the effects of targeting cellular metabolism in pancreatic cancer. Front. Physiol. 8:217, 2017.
Roy, M., and S. D. Finley. Metabolic reprogramming dynamics in tumor spheroids: Insights from a multicellular, multiscale model. PLoS Comput. Biol. 15(6):e1007053, 2019.
Schiller, J. H., D. Harrington, C. P. Belani, C. Langer, A. Sandler, J. Krook, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N. Engl. J. Med. 346(2):92–98, 2002.
Sriyudthsak, K., F. Shiraishi, and M. Y. Hirai. Mathematical modeling and dynamic simulation of metabolic reaction systems using metabolome time series data. Front. Mol. Biosci. 3:15, 2016.
Tian, Y., Z. Wang, X. Liu, J. Duan, G. Feng, Y. Yin, et al. Prediction of chemotherapeutic efficacy in non-small cell lung cancer by serum metabolomic profiling. Clin. Cancer Res. 24(9):2100–2109, 2018.
Vázquez-Gandullo, E., M. González, A. J. Ruiz-Reina, T. García-Barrena, R. A. Pérez-Grovas, J. Grávalos-Guzmán, et al. Metabolomic analysis of serum samples from patients with lung cancer. Eur. Respir. J. 44:P2702, 2014.
van de Ven, A. L., M. Wu, J. Lowengrub, S. R. McDougall, M. A. Chaplain, V. Cristini, et al. Integrated intravital microscopy and mathematical modeling to optimize nanotherapeutics delivery to tumors. AIP Adv. 2(1):11208, 2012.
Ware, M. J., L. T. Curtis, M. Wu, J. C. Ho, S. J. Corr, S. A. Curley, et al. Pancreatic adenocarcinoma response to chemotherapy enhanced with non-invasive radio frequency evaluated via an integrated experimental/computational approach. Sci. Rep. 7(1):3437, 2017.
Wen, T., L. Gao, Z. Wen, C. Wu, C. S. Tan, W. Z. Toh, et al. Exploratory investigation of plasma metabolomics in human lung adenocarcinoma. Mol. Biosyst. 9(9):2370–2378, 2013.
Wu, M., H. B. Frieboes, S. R. McDougall, M. A. Chaplain, V. Cristini, and J. Lowengrub. The effect of interstitial pressure on tumor growth: coupling with the blood and lymphatic vascular systems. J. Theor. Biol. 320:131–151, 2013.
Yizhak, K., B. Chaneton, E. Gottlieb, and E. Ruppin. Modeling cancer metabolism on a genome scale. Mol. Syst. Biol. 11(6):817, 2015.
Zaal, E. A., and C. R. Berkers. The influence of metabolism on drug response in cancer. Front. Oncol. 8:500, 2018.
Zappa, C., and S. A. Mousa. Non-small cell lung cancer: current treatment and future advances. Transl. Lung Cancer Res. 5(3):288–300, 2016.
Acknowledgments
DMM, VvB, HBF acknowledge support by National Cancer Institute Grant R15CA203605 (Frieboes).
Conflict of interest
The authors declare no known conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor Stefan M. Duma oversaw the review of this article.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Miller, H.A., Miller, D.M., van Berkel, V.H. et al. Evaluation of Lung Cancer Patient Response to First-Line Chemotherapy by Integration of Tumor Core Biopsy Metabolomics with Multiscale Modeling. Ann Biomed Eng 51, 820–832 (2023). https://doi.org/10.1007/s10439-022-03096-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10439-022-03096-8