Abstract
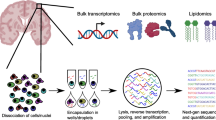
Here we demonstrate a technique to generate proteomic signatures of specific cell types within heterogeneous populations. While our method is broadly applicable across biological systems, we have limited the current work to study neural cell types isolated from human, post-mortem Alzheimer’s disease (AD) and aged-matched non-symptomatic (NS) brains. Motivating the need for this tool, we conducted an initial meta-analysis of current, human AD proteomics studies. While the results broadly corroborated major neurodegenerative disease hypotheses, cell type-specific predictions were limited. By adapting our Formaldehyde-fixed Intracellular Target-Sorted Antigen Retrieval (FITSAR) method for proteomics and applying this technique to characterize AD and NS brains, we generated enriched neuron and astrocyte proteomic profiles for a sample set of donors (available at www.fitsarpro.appspot.com). Results showed the feasibility for using FITSAR to evaluate cell-type specific hypotheses. Our overall methodological approach provides an accessible platform to determine protein presence in specific cell types and emphasizes the need for protein-compatible techniques to resolve systems complicated by cellular heterogeneity.





Similar content being viewed by others
Abbreviations
- Aβ:
-
Amyloid-beta
- AD:
-
Alzheimer’s disease
- D:
-
Donor
- DAPI:
-
4′,6-diamidino-2-phenylindole
- ECM:
-
Extracellular matrix
- ER:
-
Endoplasmic reticulum
- F:
-
Female
- FACS:
-
Fluorescence-activated cell sorting
- FSC:
-
Forward scatter
- FITSAR:
-
Formaldehyde-fixed Intracellular Target-Sorted Antigen Retrieval
- GFAP:
-
Glial fibrillary acidic protein
- H:
-
Hallmark gene sets
- K:
-
KEGG gene sets
- M:
-
Male
- NS:
-
Non-symptomatic
- O1:
-
Oligodendrocyte marker 1
- PBS:
-
Phosphate saline buffer
- PFA:
-
Paraformaldehyde
- PM:
-
Plasma membrane
- PPI:
-
Protease and phosphatase inhibitor
- R:
-
Reactome gene sets
- R#:
-
Region #
- SDS:
-
Sodium dodecyl sulfate
- SSC:
-
Side scatter
- Tryp.:
-
Trypsin
- TUBB3:
-
β-III tubulin
- y.o.:
-
Years old
References
Bard, F., et al. Peripherally administered antibodies against amyloid β-peptide enter the central nervous system and reduce pathology in a mouse model of alzheimer disease. Nat. Med. 6:916–919, 2000.
Bertram, L., and R. E. Tanzi. Genome-wide association studies in alzheimer’s disease. Hum. Mol. Genet. 18:R137–145, 2009.
Boisvert, M. M., G. A. Erikson, M. N. Shokhirev, and N. J. Allen. The aging astrocyte transcriptome from multiple regions of the mouse brain. Cell Rep. 22:269–285, 2018.
Braak, H., I. Alafuzoff, T. Arzberger, H. Kretzschmar, and K. Del Tredici. Staging of alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol. 112:389–404, 2006.
Brunk, U. T., and A. Terman. Lipofuscin: Mechanisms of age-related accumulation and influence on cell function. Free Radic. Biol. Med. 33:611–619, 2002.
Budnik, B., E. Levy, G. Harmange, and N. Slavov. Scope-ms: Mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biol. 19:161, 2018.
Cahoy, J. D., et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 28:264–278, 2008.
Chung, W. S., et al. Astrocytes mediate synapse elimination through megf10 and mertk pathways. Nature 504:394–400, 2013.
Clarke, L. E., et al. Normal aging induces a1-like astrocyte reactivity. Proc. Natl. Acad. Sci. USA 115:E1896–E1905, 2018.
Cuyvers, E., and K. Sleegers. Genetic variations underlying Alzheimer’s disease: evidence from genome-wide association studies and beyond. Lancet Neurol. 15:857–868, 2016.
Darmanis, S., et al. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. USA 112:7285–7290, 2015.
de Sousa Abreu, R., L. O. Penalva, E. M. Marcotte, and C. Vogel. Global signatures of protein and mrna expression levels. Mol. Biosyst. 5:1512–1526, 2009.
De Strooper, B., and E. Karran. The cellular phase of alzheimer’s disease. Cell 164:603–615, 2016.
Del-Aguila, J. L., et al. A single-nuclei rna sequencing study of mendelian and sporadic ad in the human brain. Alzheimers Res. Ther. 11:71, 2019.
Duong, D. The baltimore longitudinal study on aging (blsa) study. https://www.synapse.org/#!Synapse:syn3606086, 2015.
Ginsberg, S. D., S. Che, S. E. Counts, and E. J. Mufson. Single cell gene expression profiling alzheimer’s disease. NeuroRx 3:302–318, 2006.
Giri, M., M. Zhang, and Y. Lu. Genes associated with alzheimer’s disease: An overview and current status. Clin. Interv. Aging. 11:665–681, 2016.
Goeman, J. J., S. A. van de Geer, and H. C. van Houwelingen. Testing against a high dimensional alternativ. J. R. Stat. Soc. B 68:477–493, 2006.
Goltsev, Y., et al. Deep profiling of mouse splenic architecture with codex multiplexed imaging. Cell 174:968–981, 2018.
Grubman, A., et al. A single-cell atlas of entorhinal cortex from individuals with Alzheimer’s disease reveals cell-type-specific gene expression regulation. Nat. Neurosci. 22:2087–2097, 2019.
Gry, M., et al. Correlations between rna and protein expression profiles in 23 human cell lines. BMC Genom. 10:365, 2009.
Guttenplan, K. A., and S. A. Liddelow. Astrocytes and microglia: models and tools. J. Exp. Med. 216:71–83, 2018.
Hanzelmann, S., R. Castelo, and J. Guinney. Gsva: gene set variation analysis for microarray and rna-seq data. BMC Bioinform. 14:7, 2013.
Hyman, B. T., et al. National institute on aging–alzheimer’s association guidelines for the neuropathologic assessment of alzheimer’s disease. Alzheimers Dement. 8:1–13, 2012.
Jansen, I. E., et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 51:404–413, 2019.
Kita, R. Dataspectra2. https://github.com/rkita/dataspectra2, 2018.
Korin, B., et al. High-dimensional, single-cell characterization of the brain’s immune compartment. Nat. Neurosci. 20:1300–1309, 2017.
Kress, B. T., et al. Impairment of paravascular clearance pathways in the aging brain. Ann. Neurol. 76:845–861, 2014.
Lambert, J. C., et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 45:1452–1458, 2013.
Liddelow, S. A., and B. A. Barres. Reactive astrocytes: production, function, and therapeutic potential. Immunity 46:957–967, 2017.
Liddelow, S. A., et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541:481–487, 2017.
Magdeldin, S., and T. Yamamoto. Toward deciphering proteomes of formalin-fixed paraffin-embedded (ffpe) tissues. Proteomics 12:1045–1058, 2012.
Mathys, H., et al. Single-cell transcriptomic analysis of alzheimer’s disease. Nature 570:332–337, 2019.
Mattsson, N., J. M. Schott, J. Hardy, M. R. Turner, and H. Zetterberg. Selective vulnerability in neurodegeneration: Insights from clinical variants of Alzheimer’s disease. J. Neurol. Neurosurg. Psychiatry 87:1000–1004, 2016.
Metsalu, T., and J. Vilo. Clustvis: a web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 43:W566–570, 2015.
Peters, M. The banner sun health research institute (banner) study. https://www.synapse.org/#!Synapse:syn7170616, 2016.
Rothhammer, V., et al. Microglial control of astrocytes in response to microbial metabolites. Nature 557:724–728, 2018.
Sadick, J. S., M. E. Boutin, D. Hoffman-Kim, and E. M. Darling. Protein characterization of intracellular target-sorted, formalin-fixed cell subpopulations. Sci. Rep. 6:33999, 2016.
Sadick, J. S., and E. M. Darling. Processing fixed and stored adipose-derived stem cells for quantitative protein array assays. Biotechniques 63:275–280, 2017.
Seyfried, N. T., et al. A multi-network approach identifies protein-specific co-expression in asymptomatic and symptomatic Alzheimer’s disease. Cell Syst. 4:60–72, 2017.
Shen, L., and J. Jia. An overview of genome-wide association studies in Alzheimer’s disease. Neurosci. Bull. 32:183–190, 2016.
Shi, Y., et al. Apoe4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature 549:523–527, 2017.
Subramanian, A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550, 2005.
The perelman school of medicine upenn proteomics pilot (upppilot) study. https://www.synapse.org/#!Synapse:syn5477237, 2015.
Vogel, C., and E. M. Marcotte. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 13:227–232, 2012.
Weller, R. O., M. Subash, S. D. Preston, I. Mazanti, and R. O. Carare. Perivascular drainage of amyloid-beta peptides from the brain and its failure in cerebral amyloid angiopathy and Alzheimer’s disease. Brain Pathol. 18:253–266, 2008.
Yun, S. P., et al. Block of a1 astrocyte conversion by microglia is neuroprotective in models of Parkinson’s disease. Nat. Med. 24:931–938, 2018.
Zamanian, J. L., et al. Genomic analysis of reactive astrogliosis. J. Neurosci. 32:6391–6410, 2012.
Zhou, Y., et al. Human and mouse single-nucleus transcriptomics reveal trem2-dependent and trem2-independent cellular responses in Alzheimer’s disease. Nat. Med. 26:131–142, 2020.
Zhu, Y., et al. Nanodroplet processing platform for deep and quantitative proteome profiling of 10–100 mammalian cells. Nat. Commun. 9:882, 2018.
Acknowledgments
The authors would like to thank Rebecca Hamelin and Brown’s Animal Care Facility for providing sentinel rats for method optimization, Dr. Edward Stopa, Terra D. Velilla, and the Brain Tissue Resource Center for providing primary human brain samples, Mark Dooner and the COBRE Flow Cytometry Core at Rhode Island Hospital for his assistance with FACS runs, Dr. TuKiet Lam, Jean Kanyo, Wei Wei Wang, and Keck Mass Spectrometry & Proteomics Resource Core at Yale University for the mass spectrometry sample preparation and data analysis, Dr. Thomas Neubert for his feedback on the proteomics analyses, Dr. Thomas Wisniewski, Dr. Arline Faustin, and the New York University Alzheimer’s Disease Center (funded in part by PHS Grant P30 AG08051) for providing primary human brain tissue sections, and Dr. Ryosuke Kita for his help on using data spectra.
Funding
This work was supported by the National Institutes of Health (R01 AR063642 to EMD, T32 to JSS via T32 AG052909 (Wisniewski, Scharfman)), the National Science Foundation (CAREER CBET 1253189 and EAGER CBET 1547819 to EMD, GRFP 2014183678 to JSS), the Cure Alzheimer’s Fund (SAL), and the Alzheimer’s Disease Resource Center at NYU Langone (SAL and JSS).
Author Contributions
JSS, LAC, HCC, CF, SAL, and EMD designed the study and wrote the manuscript. JSS conducted all experimental work, including cell isolation, FACS, WBs, and proteomics experiments. JSS and LAC conducted all bioinformatics analyses. JSS and HCC conducted all confocal imaging.
Conflict of interest
No benefits in any form have been or will be received from a commercial party related directly or indirectly to the subject of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor Stefan M. Duma oversaw the review of this article.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sadick, J.S., Crawford, L.A., Cramer, H.C. et al. Generating Cell Type-Specific Protein Signatures from Non-symptomatic and Diseased Tissues. Ann Biomed Eng 48, 2218–2232 (2020). https://doi.org/10.1007/s10439-020-02507-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10439-020-02507-y