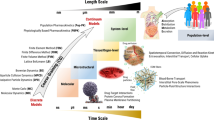
Abstract
A wide range of length and time scales are relevant to pharmacology, especially in drug development, drug design and drug delivery. Therefore, multiscale computational modeling and simulation methods and paradigms that advance the linkage of phenomena occurring at these multiple scales have become increasingly important. Multiscale approaches present in silico opportunities to advance laboratory research to bedside clinical applications in pharmaceuticals research. This is achievable through the capability of modeling to reveal phenomena occurring across multiple spatial and temporal scales, which are not otherwise readily accessible to experimentation. The resultant models, when validated, are capable of making testable predictions to guide drug design and delivery. In this review we describe the goals, methods, and opportunities of multiscale modeling in drug design and development. We demonstrate the impact of multiple scales of modeling in this field. We indicate the common mathematical and computational techniques employed for multiscale modeling approaches used in pharmacometric and systems pharmacology models in drug development and present several examples illustrating the current state-of-the-art models for (1) excitable systems and applications in cardiac disease; (2) stem cell driven complex biosystems; (3) nanoparticle delivery, with applications to angiogenesis and cancer therapy; (4) host-pathogen interactions and their use in metabolic disorders, inflammation and sepsis; and (5) computer-aided design of nanomedical systems. We conclude with a focus on barriers to successful clinical translation of drug development, drug design and drug delivery multiscale models.












Similar content being viewed by others
Abbreviations
- ABM:
-
Agent-based model
- AMD:
-
AMD3100
- BMSC:
-
Bone marrow stromal cell
- BSV:
-
Between-subject variability
- BZM:
-
Bortezomib
- CAD:
-
Computer-aided design
- CG:
-
Coarse-grained
- CXCR4:
-
C-X-C chemokine receptor type 4
- DPD:
-
Dissipative particle dynamics
- ECM:
-
Extracellular matrix
- EPR:
-
Enhanced permeability and retention
- HPV:
-
Human papillomavirus
- IFP:
-
Interstitial fluid pressure
- MC:
-
Monte Carlo
- MCMC:
-
Markov chain Monte Carlo
- MD:
-
Molecular dynamics
- MF:
-
Multiscale factorization
- MIC:
-
Myeloma initiating cell
- MM:
-
Multiple myeloma
- NC:
-
Nanocarrier
- NP:
-
Nanoparticle
- NS:
-
Navier–Stokes
- PBPK:
-
Physiologically-based pharmacokinetic
- PC:
-
Cancer progenitor cell
- PDB:
-
Protein Data Bank
- QM/MM:
-
Quantum mechanics/molecular mechanics
- RUV:
-
Residual unknown variability
- SCB:
-
Systems chemical biology
- SDF1:
-
Stromal cell-derived factor 1
- TMM:
-
Terminal multiple myeloma cell
- WHAM:
-
Weighted histogram analysis method
References
Abi Mansour, A. A., Sereda, Y. V., Yang, J. & Ortoleva, P. J. Prospective on multiscale simulation of virus-like particles: Application to computer-aided vaccine design. Vaccine, 2015. doi:10.1016/j.vaccine.2015.05.099.
Abi Mansour, A., and P. J. Ortoleva. Multiscale factorization method for simulating mesoscopic systems with atomic precision. J. Chem. Theory Comput. 10:518–523, 2014. doi:10.1021/ct400615a.
Adra, S., T. Sun, S. MacNeil, M. Holcombe, and R. Smallwood. Development of a three dimensional multiscale computational model of the human epidermis. PLoS ONE 5:e8511, 2010. doi:10.1371/journal.pone.0008511.
An, G. Agent-based computer simulation and SIRS: Building a bridge between basic science and clinical trials. Shock 16:266–273, 2001. doi:10.1097/00024382-200116040-00006.
An, G. In silico experiments of existing and hypothetical cytokine-directed clinical trials using agent-based modeling. Crit. Care Med. 32:2050–2060, 2004.
An, G., J. Bartels, and Y. Vodovotz. In silico augmentation of the drug development pipeline: examples from the study of acute inflammation. Drug Dev. Res. 72:187–200, 2011. doi:10.1002/ddr.20415.
An, G., and S. Christley. Addressing the translational dilemma: dynamic knowledge representation of inflammation using agent-based modeling. Crit. Rev. Biomed. Eng. 40:323–340, 2012.
An, G. Introduction of a framework for dynamic knowledge representation of the control structure of transplant immunology: employing the power of abstraction with a Solid Organ Transplant Agent-based Model (SOTABM). Front. Immunol. 6, 2015. doi:10.3389/fimmu.2015.00561.
Arap, W., R. Pasqualini, and E. Ruoslahti. Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science 279:377–380, 1998.
Baker, C. M., V. M. Anisimov, and A. D. MacKerell. Development of CHARMM polarizable force field for nucleic acid bases based on the classical drude oscillator model. J. Phys. Chem. B 115:580–596, 2010. doi:10.1021/jp1092338.
Beal, S. L., and L. B. Sheiner. Estimating population-kinetics. Crc. Crit. Rev. Biomed. Eng. 8:195–222, 1982.
Brennan, T., M. Fink, and B. Rodriguez. Multiscale modelling of drug-induced effects on cardiac electrophysiological activity. Eur. J. Pharm. Sci. 36:62–77, 2009. doi:10.1016/j.ejps.2008.09.013.
Britton, O. J., et al. Experimentally calibrated population of models predicts and explains intersubject variability in cardiac cellular electrophysiology. Proc. Natl. Acad. Sci. USA 110:E2098–E2105, 2013. doi:10.1073/pnas.1304382110.
Brown, J. B., and Y. Okuno. Systems biology and systems chemistry: new directions for drug discovery. Chem. Biol. 19:23–28, 2012. doi:10.1016/j.chembiol.2011.12.012.
Brown, D. R., et al. The humoral response to Gardasil® over four years as defined by Total IgG and competitive Luminex immunoassay. Hum. Vaccines 7:230–238, 2011. doi:10.4161/hv.7.2.13948.
Carpenter, T. S., E. Y. Lau, and F. C. Lightstone. Identification of a possible secondary picrotoxin-binding site on the GABAA receptor. Chem. Res. Toxicol. 26:1444–1454, 2013. doi:10.1021/tx400167b.
Champion, J. A., and S. Mitragotri. Role of target geometry in phagocytosis. Proc. Natl. Acad. Sci. USA 103:4930–4934, 2006.
Chauhan, V. P., et al. Normalization of tumour blood vessels improves the delivery of nanomedicines in a size-dependent manner. Nat. Nanotechnol. 7:383–388, 2012. doi:10.1038/nnano.2012.45.
Christley, S., C. Cockrell, and G. An. Computational studies of the intestinal host-microbiota interactome. Computation 3:2079–3197, 2015. doi:10.3390/computation3010002.
Clancy, C. E., Z. I. Zhu, and Y. Rudy. Pharmacogenetics and anti-arrhythmic drug therapy: a theoretical investigation. Am. J. Physiol. Heart Circ. Physiol. 292:H66–H75, 2007. doi:10.1152/ajpheart.00312.2006.
Clegg, L. E., and F. Mac Gabhann. Molecular mechanism matters: Benefits of mechanistic computational models for drug development. Pharmacol. Res. 99:149–154, 2015. doi:10.1016/j.phrs.2015.06.002.
Clermont, G., et al. In silico design of clinical trials: a method coming of age. Crit. Care Med. 32:2061–2070, 2004.
Collins, T. A., et al. Modeling and simulation approaches for cardiovascular function and their role in safety assessment. CPT Pharmacomet. Syst. Pharmacol. 4:e00018, 2015. doi:10.1002/psp4.18.
Csete, M., and J. Doyle. Bow ties, metabolism and disease. Trends Biotechnol. 22:446–450, 2004. doi:10.1016/j.tibtech.2004.07.007.
Davis, I. W., and D. Baker. RosettaLigand docking with full ligand and receptor flexibility. J. Mol. Biol. 385:381–392, 2009. doi:10.1016/j.jmb.2008.11.010.
Davis, M. E., Z. Chen, and D. M. Shin. Nanoparticle therapeutics: an emerging treatment modality for cancer. Nat. Rev. Drug. Discov. 7:771–782, 2008.
Decuzzi, P., and M. Ferrari. The adhesive strength of non-spherical particles mediated by specific interactions. Biomaterials 27:5307–5314, 2006. doi:10.1016/j.biomaterials.2006.05.024.
Decuzzi, P., S. Lee, B. Bhushan, and M. Ferrari. A theoretical model for the margination of particles within blood vessels. Ann. Biomed. Eng. 33:179–190, 2005. doi:10.1007/s10439-005-8976-5.
Decuzzi, P., S. Lee, M. Decuzzi, and M. Ferrari. Adhesion of microfabricated particles on vascular endothelium: a parametric analysis. Ann. Biomed. Eng. 32:793–802, 2004.
Deisboeck, T. S., Wang, Z., Macklin, P. & Cristini, V. Multiscale cancer modeling. Annu. Rev. Biomed. Eng. 13, 2011.
Deschuyteneer, M., et al. Molecular and structural characterization of the L1 virus-like particles that are used as vaccine antigens in Cervarix™, the AS04-adjuvanted HPV-16 and -18 cervical cancer vaccine. Hum. Vaccines 6:407–419, 2010.
Di Veroli, G. Y., M. R. Davies, H. Zhang, N. Abi-Gerges, and M. R. Boyett. High-throughput screening of drug-binding dynamics to HERG improves early drug safety assessment. Am. J. Physiol. Heart Circ. Physiol. 304:H104–H117, 2013. doi:10.1152/ajpheart.00511.2012.
Doyle, J., and M. Csete. Motifs, control, and stability. PLoS Biol. 3:e392, 2005. doi:10.1371/journal.pbio.0030392.
Doyle, J. C., and M. Csete. Architecture, constraints, and behavior. Proc. Natl. Acad. Sci. USA 108(Suppl 3):15624–15630, 2011. doi:10.1073/pnas.1103557108.
Dux-Santoy, L., R. Sebastian, J. Felix-Rodriguez, J. M. Ferrero, and J. Saiz. Interaction of specialized cardiac conduction system with antiarrhythmic drugs: a simulation study. IEEE Trans. Bio-med. Eng. 58:3475–3478, 2011. doi:10.1109/TBME.2011.2165213.
Fang, J., H. Nakamura, and H. Maeda. The EPR effect: unique features of tumor blood vessels for drug delivery, factors involved, and limitations and augmentation of the effect. Adv. Drug Deliv. Rev. 63:136–151, 2011.
Fang, X., A. Wallqvist, and J. Reifman. A systems biology framework for modeling metabolic enzyme inhibition of Mycobacterium tuberculosis. BMC Syst. Biol. 3:92, 2009. doi:10.1186/1752-0509-3-92.
Fang, X., A. Wallqvist, and J. Reifman. Modeling synergistic drug inhibition of Mycobacterium tuberculosis growth in murine macrophages. Mol. BioSyst. 7:2622–2636, 2011. doi:10.1039/c1mb05106g.
Finley, S. D., P. Angelikopoulos, P. Koumoutsakos, and A. S. Popel. Pharmacokinetics of anti-VEGF agent aflibercept in cancer predicted by data driven, molecular-detailed model. CPT Pharmacomet. Syst. Pharmacol. 4(11):641–649, 2015.
Finley, S. D., L. H. Chu, and A. S. Popel. Computational systems biology approaches to anti-angiogenic cancer therapeutics. Drug Discov. Today 20:187–197, 2015. doi:10.1016/j.drudis.2014.09.026.
Fogelson, A. L., and R. D. Guy. Immersed-boundary-type models of intravascular platelet aggregation. Comput. Method Appl. M 197:2087–2104, 2008. doi:10.1016/j.cma.2007.06.030.
Garfinkel, A., et al. Preventing ventricular fibrillation by flattening cardiac restitution. Proc. Natl. Acad. Sci. USA 97:6061–6066, 2000. doi:10.1073/pnas.090492697.
Geerts, H., A. Spiros, P. Roberts, and R. Carr. Has the time come for predictive computer modeling in CNS drug discovery and development? CPT Pharmacomet. Syst. Pharmacol. 1:e16, 2012. doi:10.1038/psp.2012.17.
Geng, Y., et al. Shape effects of filaments versus spherical particles in flow and drug delivery. Nat. Nanotechnol. 2:249–255, 2007.
Grosch, J., J. Yang, A. Shen, Y. V. Sereda, and P. Ortoleva. Broad spectrum assessment of the epitope fluctuation—immunogenicity hypothesis. Vaccine 2015. doi:10.1016/j.vaccine.2015.06.111.
Haddish-Berhane, N., J. L. Rickus, and K. Haghighi. The role of multiscale computational approaches for rational design of conventional and nanoparticle oral drug delivery systems. Int. J. Nanomed. 2:315, 2007.
Hall, B. Lie Groups, Lie Algebras, and Representations: An Elementary Introduction. Berlin: Springer, 2004.
Haun, J. B., and D. A. Hammer. Quantifying nanoparticle adhesion mediated by specific molecular interactions. Langmuir 24:8821–8832, 2008.
Heldin, C.-H., K. Rubin, K. Pietras, and A. Ostman. High interstitial fluid pressure [mdash] an obstacle in cancer therapy. Nat. Rev. Cancer 4:806–813, 2004.
Holford, N. H. G., H. C. Kimko, J. P. R. Monteleone, and C. C. Peck. Simulation of clinical trials. Annu. Rev. Pharmacol. 40:209–234, 2000. doi:10.1146/Annurev.Pharmtox.40.1.209.
Hossain, S. S., et al. In silico vascular modeling for personalized nanoparticle delivery. Nanomedicine 8:343–357, 2013.
Hunt, C. A., R. C. Kennedy, S. H. Kim, and G. E. Ropella. Agent-based modeling: a systematic assessment of use cases and requirements for enhancing pharmaceutical research and development productivity. Wiley Interdiscip Rev Syst Biol Med 5:461–480, 2013. doi:10.1002/wsbm.1222.
Hyakutake, T., and S. Nagai. Numerical simulation of red blood cell distributions in three-dimensional microvascular bifurcations. Microvasc. Res. 97:115–123, 2015. doi:10.1016/j.mvr.2014.10.001.
Jain, R. K. Barriers to drug delivery in solid tumors. Sci. Am. 58–65:e29528, 1994.
Joshi, H., S. Cheluvaraja, E. Somogyi, D. R. Brown, and P. J. Ortoleva. A molecular dynamics study of loop fluctuation in human papillomavirus type 16 virus-like particles: a possible indicator of immunogenicity. Vaccine 29:9423–9430, 2011. doi:10.1016/j.vaccine.2011.10.039.
Kakde, D., D. Jain, V. Shrivastava, R. Kakde, and A. Patil. Cancer therapeutics-opportunities, challenges and advances in drug delivery. J. Appl. Pharm. Sci. 1(9):1–10, 2011.
Kim, M., R. J. Gillies, and K. A. Rejniak. Current advances in mathematical modeling of anti-cancer drug penetration into tumor tissues. Front. Oncol. 3:120, 2013.
King, M. R., and D. A. Hammer. Multiparticle adhesive dynamics: hydrodynamic recruitment of rolling leukocytes. Proc. Natl. Acad. Sci. USA 98:14919–14924, 2001. doi:10.1073/pnas.261272498.
King, M. R., S. D. Rodgers, and D. A. Hammer. Hydrodynamic collisions suppress fluctuations in the rolling velocity of adhesive blood cells. Langmuir 17:4139–4143, 2001. doi:10.1021/La010234b.
Kumar, R., G. Clermont, Y. Vodovotz, and C. C. Chow. The dynamics of acute inflammation. J. Theor. Biol. 230:145–155, 2004. doi:10.1016/j.jtbi.2004.04.044.
Lalonde, R. L., et al. Model-based drug development. Clin. Pharmacol. Ther. 82:21–32, 2007. doi:10.1038/sj.clpt.6100235.
Li, X. L., K. L. Yao, and Z. L. Liu. CFD study on the magnetic fluid delivering in the vessel in high-gradient magnetic field. J. Magn. Magn. Mater. 320:1753–1758, 2008. doi:10.1016/j.jmmm.2008.01.041.
Liu, J., R. Bradley, D. M. Eckmann, P. S. Ayyaswamy, and R. Radhakrishnan. Multiscale modeling of functionalized nanocarriers in targeted drug delivery. Curr. Nanosci. 7:727, 2011.
Liu, J., et al. Computational model for nanocarrier binding to endothelium validated using in vivo, in vitro, and atomic force microscopy experiments. Proc. Natl. Acad. Sci. 107:16530–16535, 2010. doi:10.1073/pnas.1006611107.
Lowy, D. R., and J. T. Schiller. Prophylactic human papillomavirus vaccines. J. Clin. Invest. 116:1167–1173, 2006. doi:10.1172/JCI28607.
Lu, D., et al. Model-based meta-analysis for quantifying Paclitaxel dose response in cancer patients. CPT Pharmacomet. Syst. Pharmacol. 3:e115, 2014. doi:10.1038/psp.2014.14.
Mahmoudi, M., et al. Multiphysics flow modeling and in vitro toxicity of iron oxide nanoparticles coated with poly(vinyl alcohol). J. Phys. Chem. C 113:2322–2331, 2009. doi:10.1021/jp809453v.
Malisi, C., et al. Binding pocket optimization by computational protein design. PLoS ONE 7:e52505, 2012. doi:10.1371/journal.pone.0052505.
Mandell, D. J., E. A. Coutsias, and T. Kortemme. Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat. Methods 6:551–552, 2009.
May, E. E., A. Leitao, A. Tropsha, and T. I. Oprea. A systems chemical biology study of malate synthase and isocitrate lyase inhibition in Mycobacterium tuberculosis during active and NRP growth. Comput. Biol. Chem. 47:167–180, 2013. doi:10.1016/j.compbiolchem.2013.07.002.
May, E., et al. Understanding virulence mechanisms in M. tuberculosis infection via a circuit-based simulation framework. Conf. Proc. IEEE Eng. Med. Biol. Soc. 4953–4955:2008, 2008. doi:10.1109/IEMBS.2008.4650325.
Meiler, J., and D. Baker. ROSETTALIGAND: protein-small molecule docking with full side-chain flexibility. Proteins 65:538–548, 2006. doi:10.1002/prot.21086.
Mi, Q., B. Riviere, G. Clermont, D. L. Steed, and Y. Vodovotz. Agent-based model of inflammation and wound healing: insights into diabetic foot ulcer pathology and the role of transforming growth factor-beta1. Wound Repair Regen. 15:671–682, 2007. doi:10.1111/j.1524-475X.2007.00271.x.
Milligan, P. A., et al. Model-based drug development: a rational approach to efficiently accelerate drug development. Clin. Pharmacol. Ther. 93:502–514, 2013. doi:10.1038/clpt.2013.54.
Modok, S., et al. Transport kinetics of four-coordinate and six-coordinate platinum compounds in the multicell layer tumour model. Br. J. Cancer 97:194–200, 2007. doi:10.1038/sj.bjc.6603854.
Moreno, J. D., et al. A computational model to predict the effects of class I anti-arrhythmic drugs on ventricular rhythms. Sci. Trans. Med. 3:98ra83, 2011. doi:10.1126/scitranslmed.3002588.
Moreno, J. D., et al. Ranolazine for congenital and acquired late INa-linked arrhythmias: in silico pharmacological screening. Circ. Res. 113:e50–e61, 2013. doi:10.1161/CIRCRESAHA.113.301971.
Mould, D. R. Model-based meta-analysis: an important tool for making quantitative decisions during drug development. Clin. Pharmacol. Ther. 92:283–286, 2012. doi:10.1038/clpt.2012.122.
Mould, D. R. Models for disease progression: new approaches and uses. Clin. Pharmacol. Ther. 92:125–131, 2012. doi:10.1038/clpt.2012.53.
Muller, K., Fedosov, D. A. & Gompper, G. Margination of micro- and nano-particles in blood flow and its effect on drug delivery. Sci. Rep. 4: doi:10.1038/srep04871. http://www.nature.com/srep/2014/140502/srep04871/abs/srep04871.html—supplementary-information, 2014.
Nakamura, H., et al. progesterone regulates cardiac repolarization through a nongenomic pathway: an in vitro patch-clamp and computational modeling study. Circulation 116:2913–2922, 2007. doi:10.1161/circulationaha.107.702407.
Obiol-Pardo, C., J. Gomis-Tena, F. Sanz, J. Saiz, and M. Pastor. A multiscale simulation system for the prediction of drug-induced cardiotoxicity. J. Chem. Inf. Model. 51:483–492, 2011. doi:10.1021/ci100423z.
Oprea, T. I., E. E. May, A. Leitao, and A. Tropsha. Computational systems chemical biology. Methods Mol. Biol. 672:459–488, 2011. doi:10.1007/978-1-60761-839-3_18.
Oprea, T. I., A. Tropsha, J. L. Faulon, and M. D. Rintoul. Systems chemical biology. Nat. Chem. Biol. 3:447–450, 2007. doi:10.1038/nchembio0807-447.
Osborne, J. M., et al. A hybrid approach to multi-scale modelling of cancer. Philos. Trans. A Math. Phys. Eng. Sci. 368:5013–5028, 2010.
Pozrikidis, C. Numerical simulation of blood and interstitial flow through a solid tumor. J. Math. Biol. 60:75–94, 2010.
Preliminary report: effect of encainide and flecainide on mortality in a randomized trial of arrhythmia suppression after myocardial infarction. The Cardiac Arrhythmia Suppression Trial (CAST) Investigators. N. Engl. J. Med. 321:406–412, 1989.
Qu, Z., J. N. Weiss, and A. Garfinkel. Cardiac electrical restitution properties and stability of reentrant spiral waves: a simulation study. Am. J. Physiol. 276:H269–H283, 1999.
Recanatini, M., A. Cavalli, and M. Masetti. Modeling HERG and its interactions with drugs: recent advances in light of current potassium channel simulations. ChemMedChem 3:523–535, 2008. doi:10.1002/cmdc.200700264.
Remien, C. H., F. R. Adler, L. Waddoups, T. D. Box, and N. L. Sussman. Mathematical modeling of liver injury and dysfunction after acetaminophen overdose: Early discrimination between survival and death. Hepatology 56:727–734, 2012. doi:10.1002/hep.25656.
Sager, J. E., J. Yu, I. Ragueneau-Majlessi, and N. Isoherranen. Physiologically based pharmacokinetic (PBPK) modeling and simulation approaches: a systematic review of published models, applications and model verification. Drug Metab. Dispos. 2015. doi:10.1124/dmd.115.065920.
Sarkar, A. X., and E. A. Sobie. Quantification of repolarization reserve to understand interpatient variability in the response to proarrhythmic drugs: a computational analysis. Heart Rhythm. 8:1749–1755, 2011. doi:10.1016/j.hrthm.2011.05.023.
Shah, D. K., N. Haddish-Berhane, and A. Betts. Bench to bedside translation of antibody drug conjugates using a multiscale mechanistic PK/PD model: a case study with brentuximab-vedotin. J. Pharmacokinet Pharmacodyn. 39:643–659, 2012.
Shipley, R. J., and S. J. Chapman. Multiscale modelling of fluid and drug transport in vascular tumours. Bull. Math. Biol. 72:1464–1491, 2010. doi:10.1007/s11538-010-9504-9.
Siegel, R. L., K. D. Miller, and A. Jemal. Cancer statistics, 2015. CA Cancer J. Clin. 65:5–29, 2015. doi:10.3322/caac.21254.
Singharoy, A., H. Joshi, Y. Miao, and P. J. Ortoleva. Space warping order parameters and symmetry: application to multiscale simulation of macromolecular assemblies. J. Phys. Chem. B 116:8423–8434, 2012. doi:10.1021/jp2119247.
Soding, J. Protein homology detection by HMM-HMM comparison. Bioinformatics (Oxford, England) 21:951–960, 2005.
Sohrabi, S., J. Zheng, E. A. Finol, and Y. Liu. Numerical simulation of particle transport and deposition in the pulmonary vasculature. J. Biomech. Eng. 136:121010, 2014.
Song, H.-S., W. Cannon, A. Beliaev, and A. Konopka. Mathematical modeling of microbial community dynamics: a methodological review. Processes 2:711, 2014.
Sorger, P. K. et al. Quantitative and systems pharmacology in the post-genomic era: new approaches to discovering drugs and understanding therapeutic mechanisms. An NIH White Paper by the QSP Workshop Group. (2011). https://www.nigms.nih.gov/training/documents/systemspharmawpsorger2011.pdf.
Stamatakos, G. S., E. A. Kolokotroni, D. D. Dionysiou, E. Georgiadi, and C. Desmedt. An advanced discrete state-discrete event multiscale simulation model of the response of a solid tumor to chemotherapy: mimicking a clinical study. J. Theor. Biol. 266:124–139, 2010. doi:10.1016/j.jtbi.2010.05.019.
Su, J. & Henson, M. A. Circadian gating of the mammalian cell cycle restriction point: a mathematical analysis. Life Sci. Lett. IEEE:1–1, 2015, doi:10.1109/LLS.2015.2449511.
Su, J., et al. Targeting the biophysical properties of the myeloma initiating cell niches: a pharmaceutical synergism analysis using multi-scale agent-based modeling. PLoS ONE 9:e85059, 2014.
Sun, X., et al. Cytokine combination therapy prediction for bone remodeling in tissue engineering based on the intracellular signaling pathway. Biomaterials 33:8265–8276, 2012.
Szklarczyk, O. M., S. J. Bachmann, and W. F. van Gunsteren. A polarizable empirical force field for molecular dynamics simulation of liquid hydrocarbons. J. Comput. Chem. 35:789–801, 2014. doi:10.1002/jcc.23551.
Tan, J., S. Shah, A. Thomas, H. D. Ou-Yang, and Y. Liu. The influence of size, shape and vessel geometry on nanoparticle distribution. Microfluidics Nanofluidics 14:77–87, 2013.
Tan, J., A. Thomas, and Y. Liu. Influence of red blood cells on nanoparticle targeted delivery in microcirculation. Soft. Matter. 8(6):1934–1946, 2012.
Tan, J., S. Wang, J. Yang, and Y. Liu. Coupled particulate and continuum model for nanoparticle targeted delivery. Comput. Struct. 122:128–134, 2013.
Tang, L., et al. Computational modeling of 3D tumor growth and angiogenesis for chemotherapy evaluation. PLoS ONE 9:e83962, 2014. doi:10.1371/journal.pone.0083962.
ten Tusscher, K. H. W. J., and A. V. Panfilov. Alternans and spiral breakup in a human ventricular tissue model. Am. J. Physiol. Heart Circ. Physiol. 291:H1088–H1100, 2006.
The Sicilian gambit. A new approach to the classification of antiarrhythmic drugs based on their actions on arrhythmogenic mechanisms. Task Force of the Working Group on Arrhythmias of the European Society of Cardiology. Circulation 84:1831–1851, 1991.
The ‘Sicilian Gambit’. A new approach to the classification of antiarrhythmic drugs based on their actions on arrhythmogenic mechanisms. The Task Force of the Working Group on Arrhythmias of the European Society of Cardiology. Eur. Heart J. 12:1112–1131, 1991.
Thomas, D. G., S. Jaramillo-Riveri, D. J. Baxter, and W. R. Cannon. Comparison of optimal thermodynamic models of the tricarboxylic acid cycle from heterotrophs, cyanobacteria, and green sulfur bacteria. J. Phys. Chem. B 2014. doi:10.1021/jp5075913.
Torchilin, V. Tumor delivery of macromolecular drugs based on the EPR effect. Adv. Drug Deliv. Rev. 63:131–135, 2011.
Trivedi, A., R. E. Lee, and B. Meibohm. Applications of pharmacometrics in the clinical development and pharmacotherapy of anti-infectives. Expert Rev. Clin. Phar. 6:159–170, 2013. doi:10.1586/ECP.13.6.
Trotter, H. F. On the product of semi-groups of operators. Proc. Am. Math. Soc. 10:545–551, 1959. doi:10.1090/S0002-9939-1959-0108732-6.
Uma, B., T. N. Swaminathan, R. Radhakrishnan, D. M. Eckmann, and P. S. Ayyaswamy. Nanoparticle Brownian motion and hydrodynamic interactions in the presence of flow fields. Phys Fluids 23:073602, 2011. doi:10.1063/1.3611026.
Uppal, A., S. C. Wightman, S. Ganai, R. R. Weichselbaum, and G. An. Investigation of the essential role of platelet-tumor cell interactions in metastasis progression using an agent-based model. Theor. Biol. Med. Model 11:17, 2014. doi:10.1186/1742-4682-11-17.
van der Graaf, P. H. CPT: pharmacometrics and systems pharmacology. CPT Pharmacomet. Syst. Pharmacol 1:e8, 2012. doi:10.1038/psp.2012.8.
Vanommeslaeghe, K., et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 31:671–690, 2009.
Vicini, P., and B. P. Smith. Whither pharmacometrics?: present state and future choices. Clin. Pharmacol. Ther. 95:567–571, 2014. doi:10.1038/clpt.2014.72.
Vicini, P., and P. H. van der Graaf. Systems pharmacology for drug discovery and development: paradigm shift or flash in the pan? Clin. Pharmacol. Ther. 93:379–381, 2013. doi:10.1038/clpt.2013.40.
Waldo, A. L., et al. Effect of d-sotalol on mortality in patients with left ventricular dysfunction after recent and remote myocardial infarction. The SWORD Investigators. Survival With Oral d-Sotalol. Lancet 348:7–12, 1996.
Wang, C., P. Bradley, and D. Baker. Protein-protein docking with backbone flexibility. J. Mol. Biol. 373:503–519, 2007. doi:10.1016/j.jmb.2007.07.050.
Wang, Z., and T. S. Deisboeck. Mathematical modeling in cancer drug discovery. Drug Discov. Today 19:145–150, 2014. doi:10.1016/j.drudis.2013.06.015.
Weiss, J. N., et al. Electrical restitution and cardiac fibrillation. J. Cardiovasc. Electrophysiol. 13:292–295, 2002.
Woltosz, W. S. If we designed airplanes like we design drugsaEuro broken vertical bar. J. Comput. Aid. Mol. Des. 26:159–163, 2012. doi:10.1007/s10822-011-9490-5.
Xie, Y., L. T. Izu, D. M. Bers, and D. Sato. Arrhythmogenic transient dynamics in cardiac myocytes. Biophys. J . 106:1391–1397, 2014. doi:10.1016/j.bpj.2013.12.050.
Yang, K. & Ma, Y.-Q. Computer simulation of the translocation of nanoparticles with different shapes across a lipid bilayer. Nat. Nano 5:579–583. http://www.nature.com/nnano/journal/v5/n8/abs/nnano.2010.141.html—supplementary-information, 2010.
Yang, P. C., J. Kurokawa, T. Furukawa, and C. E. Clancy. Acute effects of sex steroid hormones on susceptibility to cardiac arrhythmias: a simulation study. PLoS Comput. Biol. 6:e1000658, 2010. doi:10.1371/journal.pcbi.1000658.
Yang, J., A. Singharoy, Y. V. Sereda, and P. J. Ortoleva. Quasiequivalence of multiscale coevolution and ensemble MD simulations: a demonstration with lactoferrin. Chem. Phys. Lett. 616–617:154–160, 2014. doi:10.1016/j.cplett.2014.10.020.
Zemzemi, N., et al. Computational assessment of drug-induced effects on the electrocardiogram: from ion channel to body surface potentials. Br. J. Pharmacol. 168:718–733, 2013. doi:10.1111/j.1476-5381.2012.02200.x.
Zeng, X., and S. Li. Multiscale modeling and simulation of soft adhesion and contact of stem cells. J. Mech. Behav. Biomed. Mater. 4:180–189, 2011. doi:10.1016/j.jmbbm.2010.06.002.
Zhang, J. Z., et al. Structure-function map of the receptor site for beta-scorpion toxins in domain II of voltage-gated sodium channels. J. Biol. Chem. 286:33641–33651, 2011. doi:10.1074/jbc.M111.282509.
Zhang, J. Z., et al. Mapping the interaction site for a beta-scorpion toxin in the pore module of domain III of voltage-gated Na(+) channels. J. Biol. Chem. 287:30719–30728, 2012. doi:10.1074/jbc.M112.370742.
Zhou, Q., G. C. Bett, and R. L. Rasmusson. Markov models of use-dependence and reverse use-dependence during the mouse cardiac action potential. PLoS ONE 7:e42295, 2012. doi:10.1371/journal.pone.0042295.
Zhu, Z. I., and C. E. Clancy. L-type Ca2+channel mutations and T-wave alternans: a model study. Am. J. Physiol. Heart Circ. Physiol. 293:H3480–H3489, 2007. doi:10.1152/ajpheart.00476.2007.
Ziraldo, C., et al. A computational, tissue-realistic model of pressure ulcer formation in individuals with spinal cord injury. PLoS Comput. Biol. 11:e1004309, 2015. doi:10.1371/journal.pcbi.1004309.
Acknowledgments
This work was supported in part by NIH grants R01CA138264 (ASP), U01HL126273 (CEC), U01EB016027 (DME), R01EB006818 (DME), R01-GM-115839 and P30-DK-42086 (GA), R01GM077138 (JPS) and R15EB015105 (YL) as well as EPA grant R835001 (JPS). WRC was funded under the Laboratory Directed Research Program at the Pacific Northwest National Laboratory. PNNL is operated by Battelle for the U.S. Department of Energy under Contract DE-AC06-76RLO.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Associate Editor Thomas Yankeelov oversaw the review of this article.
Rights and permissions
About this article
Cite this article
Clancy, C.E., An, G., Cannon, W.R. et al. Multiscale Modeling in the Clinic: Drug Design and Development. Ann Biomed Eng 44, 2591–2610 (2016). https://doi.org/10.1007/s10439-016-1563-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10439-016-1563-0