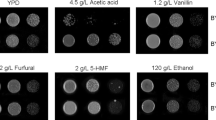
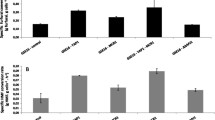
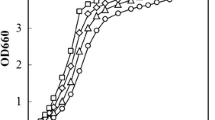
Abstract
Toxic concentrations of monocarboxylic weak acids present in lignocellulosic hydrolyzates affect cell integrity and fermentative performance of Saccharomyces cerevisiae. In this work, we report the deletion of the general catabolite repressor Mig1p as a strategy to improve the tolerance of S. cerevisiae towards inhibitory concentrations of acetic, formic or levulinic acid. In contrast with the wt yeast, where the growth and ethanol production were ceased in presence of acetic acid 5 g/L or formic acid 1.75 g/L (initial pH not adjusted), the m9 strain (Δmig1::kan) produced 4.06 ± 0.14 and 3.87 ± 0.06 g/L of ethanol, respectively. Also, m9 strain tolerated a higher concentration of 12.5 g/L acetic acid (initial pH adjusted to 4.5) without affecting its fermentative performance. Moreover, m9 strain produced 33% less acetic acid and 50–70% less glycerol in presence of weak acids, and consumed acetate and formate as carbon sources under aerobic conditions. Our results show that the deletion of Mig1p provides a single gene deletion target for improving the acid tolerance of yeast strains significantly.






Similar content being viewed by others
References
Bracey D, Holyoak C, Nebe-von Caron G, Coote P (1998) Determination of the intracellular pH (pHi) of growing cells of Saccharomyces cerevisiae: the effect of reduced-expression of the membrane H+-ATPase. J Microbiol Methods 31:113–125. https://doi.org/10.1016/S0167-7012(97)00095-X
Burgard A, Pharkya P, Osterhout R (2011) Microorganisms for the production of adipic acid and other compounds. US Patent Number: US7799545B2
Cantarella M, Cantarella L, Alberto Gallifuoco AS, Alfani F (2004) Effect of inhibitors released during steam-explosion treatment of poplar wood on subsequent enzymatic hydrolysis and SSF. Biotechnol Prog 20:200–206. https://doi.org/10.1021/bp0257978
Cao H, Yue M, Li S, Bai X, Zhao X, Du Y (2011) The impact of MIG1 and/or MIG2 disruption on aerobic metabolism of succinate dehydrogenase negative Saccharomyces cerevisiae. Appl Microbiol Biotechnol 89:733–738. https://doi.org/10.1007/s00253-010-2894-7
Casal M, Paiva S, Queirós O, Soares-Silva I (2008) Transport of carboxylic acids in yeasts. FEMS Microbiol Rev 32:974–994. https://doi.org/10.1111/j.1574-6976.2008.00128.x
Casey E, Sedlak M, Ho NWY, Mosier NS (2010) Effect of acetic acid and pH on the cofermentation of glucose and xylose to ethanol by a genetically engineered strain of Saccharomyces cerevisiae. FEMS Yeast Res 10:385–393. https://doi.org/10.1111/j.1567-1364.2010.00623.x
Crozet P, Margalha L, Confraria A, Rodrigues A, Martinho C, Adamo M, Elias CA, Baena-GonzÃlez E (2014) Mechanisms of regulation of SNF1/AMPK/SnRK1 protein kinases. Front Plant Sci 5:1–17. https://doi.org/10.3389/fpls.2014.00190
Demeke MM, Dumortier F, Li Y, Broeckx T, Foulquié-Moreno MR, Thevelein JM (2013) Combining inhibitor tolerance and D-xylose fermentation in industrial Saccharomyces cerevisiae for efficient lignocellulose-based bioethanol production. Biotechnol Biofuels 6:120. https://doi.org/10.1186/1754-6834-6-120
Dikicioglu D, Pir P, Onsan ZI, Ulgen KO, Kirdar B, Oliver SG (2008) Integration of metabolic modeling and phenotypic data in evaluation and improvement of ethanol production using respiration-deficient mutants of Saccharomyces cerevisiae. Appl Environ Microbiol 74:5809–5816. https://doi.org/10.1128/AEM.00009-08
Eraso P, Gancedo C (1987) Activation of yeast plasma membrane ATPase by acid pH during growth. FEBS Lett 224:187–192. https://doi.org/10.1016/0014-5793(87)80445-3
Fleet G (1992) Spoilage yeasts. Crit Rev Biotechnol. https://doi.org/10.3109/07388559209069186
Guerreiro JF, Mira NP, Sá-Correia I (2012) Adaptive response to acetic acid in the highly resistant yeast species Zygosaccharomyces bailii revealed by quantitative proteomics. Proteomics 12:2303–2318. https://doi.org/10.1002/pmic.201100457
Hasunuma T, Sanda T, Yamada R, Yoshimura K, Ishii J, Kondo A (2011) Metabolic pathway engineering based on metabolomics confers acetic and formic acid tolerance to a recombinant xylose-fermenting strain of Saccharomyces cerevisiae. Microb Cell Fact 10:2. https://doi.org/10.1186/1475-2859-10-2
Holyoak C, Stratford M, McMullin Z, Cole M, Crimmins K, Brown A, Coote P (1996) Activity of the plasma membrane H(+)-ATPase and optimal glycolytic flux are required for rapid adaptation and growth of Saccharomyces cerevisiae in the presence of the weak-acid preservative sorbic acid. Appl Envir Microbiol 62:3158–3164
Holyoak CD, Bracey D, Piper PW, Kuchler K, Coote PJ (1999) The Saccharomyces cerevisiae weak-acid-inducible ABC transporter Pdr12 transports fluorescein and preservative anions from the cytosol by an energy-dependent mechanism. J Bacteriol 181:4644–4652
Hou J, Scalcinati G, Oldiges M, Vemuri GN (2010) Metabolic impact of increased NADH availability in Saccharomyces cerevisiae. Appl Environ Microbiol 76:851–859. https://doi.org/10.1128/AEM.02040-09
Hyland PB, Mun SLS, Mahadevan R (2013) Prediction of weak acid toxicity in Saccharomyces cerevisiae using genome-scale metabolic models. Ind Biotechnol 9:229–235. https://doi.org/10.1089/ind.2013.0004
Imai T, Ohno T (1995) The relationship between viability and intracellular pH in the yeast Saccharomyces cerevisiae. Appl Environ Microbiol 61:3604–3608
Inaba T, Watanabe D, Yoshiyama Y, Tanaka K, Ogawa J, Takagi H, Shimoi H, Shima J (2013) An organic acid-tolerant HAA1-overexpression mutant of an industrial bioethanol strain of Saccharomyces cerevisiae and its application to the production of bioethanol from sugarcane molasses. AMB Express 3:74. https://doi.org/10.1186/2191-0855-3-74
Jönsson LJ, Martín C (2016) Pretreatment of lignocellulose: formation of inhibitory by-products and strategies for minimizing their effects. Bioresour Technol 199:103–112. https://doi.org/10.1016/j.biortech.2015.10.009
Klein CJL, Olsson L, Rønnow B, Mikkelsen JD, Nielsen J (1996) Alleviation of glucose repression of maltose metabolism by MIG1 disruption in Saccharomyces cerevisiae. Appl Environ Microbiol 62:4441–4449
Klein CJL, Olsson L, Rønnow B, Mikkelsen JD, Nielsen J (1997) Glucose and maltose metabolism in MIG1-disrupted and MAL-constitutive strains of Saccharomyces cerevisiae. Food Technol Biotechnol 35:287–292
Klein CJL, Rasmussen JJ, Rønnow B, Olsson L, Nielsen J (1999) Investigation of the impact of MIG1 and MIG2 on the physiology of Saccharomyces cerevisiae. J Biotechnol 68:197–212. https://doi.org/10.1016/S0168-1656(98)00205-3
Klinke HB, Thomsen AB, Ahring BK (2004) Inhibition of ethanol-producing yeast and bacteria by degradation products produced during pre-treatment of biomass. Appl Microbiol Biotechnol 66:10–26. https://doi.org/10.1007/s00253-004-1642-2
Kong QX, Gu JG, Cao LM, Zhang AL, Chen X, Zhao XM (2006) Improved production of ethanol by deleting FPS1 and over-expressing GLT1 in Saccharomyces cerevisiae. Biotechnol Lett 28:2033–2038. https://doi.org/10.1007/s10529-006-9185-5
Larsson S, Palmqvist E, Hahn-Hgerdal B, Tengborg C, Stenberg K, Zacchi G, Nilvebrant NO (1999) The generation of fermentation inhibitors during dilute acid hydrolysis of softwood. Enzyme Microb Technol 24:151–159. https://doi.org/10.1016/s0141-0229(98)00101-x
Li YC, Gou ZX, Liu ZS, Tang YQ, Akamatsu T, Kida K (2014) Synergistic effects of TAL1 over-expression and PHO13 deletion on the weak acid inhibition of xylose fermentation by industrial Saccharomyces cerevisiae strain. Biotechnol Lett 36:2011–2021. https://doi.org/10.1007/s10529-014-1581-7
Lindahl L, Genheden S, Eriksson LA, Olsson L, Bettiga M (2016) Sphingolipids contribute to acetic acid resistance in Zygosaccharomyces bailii. Biotechnol Bioeng 113:744–753. https://doi.org/10.1002/bit.25845
Lindberg L, Santos AXS, Riezman H, Olsson L, Bettiga M (2013) Lipidomic profiling of Saccharomyces cerevisiae and Zygosaccharomyces bailii reveals critical changes in lipid composition in response to acetic acid stress. PLoS One. https://doi.org/10.1371/journal.pone.0073936
Lu Y, Cheng YF, He XP, Guo XN, Zhang BR (2012) Improvement of robustness and ethanol production of ethanologenic Saccharomyces cerevisiae under co-stress of heat and inhibitors. J Ind Microbiol Biotechnol 39:73–80. https://doi.org/10.1007/s10295-011-1001-0
Luttik MAH, Overkamp KM, Kötter P, De Vries S, Van Dijken JP, Pronk JT (1998) The Saccharomyces cerevisiae NDE1 and NDE2 genes encode separate mitochondrial NADH dehydrogenases catalyzing the oxidation of cytosolic NADH. J Biol Chem 273:24529–24534. https://doi.org/10.1074/jbc.273.38.24529
Ma C, Wei X, Sun C, Zhang F, Xu J, Zhao X, Bai F (2015) Improvement of acetic acid tolerance of Saccharomyces cerevisiae using a zinc-finger-based artificial transcription factor and identification of novel genes involved in acetic acid tolerance. Appl Microbiol Biotechnol 99:2441–2449. https://doi.org/10.1007/s00253-014-6343-x
Van Maris AJA, Geertman JMA, Vermeulen A, Groothuizen MK, Winkler AA, Piper MDW, Van Dijken JP, Pronk JT (2004) directed evolution of pyruvate decarboxylase-negative Saccharomyces cerevisiae, yielding a C2-independent, glucose-tolerant, and pyruvate-hyperproducing yeast. Appl Environ Microbiol 70:159–166. https://doi.org/10.1128/AEM.70.1.159-166.2004
Marres CAM, Vries S, Grivell LA (1991) Isolation and inactivation of the nuclear gene encoding the rotenone-insensitive internal NADH: ubiquinone oxidoreductase of mitochondria from Saccharomyces cerevisiae. Eur J Biochem 195:857–862. https://doi.org/10.1111/j.1432-1033.1991.tb15775.x
Meijnen J-P, Randazzo P, Foulquié-Moreno MR, van den Brink J, Vandecruys P, Stojiljkovic M, Dumortier F, Zalar P, Boekhout T, Gunde-Cimerman N, Kokošar J, Štajdohar M, Curk T, Petrovič U, Thevelein JM (2016) Polygenic analysis and targeted improvement of the complex trait of high acetic acid tolerance in the yeast Saccharomyces cerevisiae. Biotechnol Biofuels 9:5. https://doi.org/10.1186/s13068-015-0421-x
Merico A, Capitanio D, Viigenti I, Ranzi B, Compagno C (2003) Aerobic sugar metabolism in the spoilage yeast. FEMS Yeast Res 4:277–283. https://doi.org/10.1016/s1567-1356(03)00167-3
Mira NP, Lourenço AB, Fernandes AR, Becker JD, Sá-Correia I (2009) The RIM101 pathway has a role in Saccharomyces cerevisiae adaptive response and resistance to propionic acid and other weak acids. FEMS Yeast Res 9:202–216. https://doi.org/10.1111/j.1567-1364.2008.00473.x
Mira NP, Palma M, Guerreiro JF, Sá-Correia I (2010) Genome-wide identification of Saccharomyces cerevisiae genes required for tolerance to acetic acid. Microb Cell Fact 9:79. https://doi.org/10.1186/1475-2859-9-79
Mira NP, Teixeira MC, Sá-Correia I (2010) Adaptive response and tolerance to weak acids in Saccharomyces cerevisiae: a genome-wide view. OMICS 14:525–540. https://doi.org/10.1089/omi.2010.0072
Mollapour M, Piper PW (2007) Hog1 mitogen-activated protein kinase phosphorylation targets the yeast Fps1 aquaglyceroporin for endocytosis, thereby rendering cells resistant to acetic acid. Mol Cell Biol 27:6446–6456. https://doi.org/10.1128/MCB.02205-06
Mollapour M, Shepherd A, Piper PW (2008) Novel stress responses facilitate Saccharomyces cerevisiae growth in the presence of the monocarboxylate preservatives. Yeast 25:169–177. https://doi.org/10.1002/yea.1576
Nissen TL, Kielland-Brandt MC, Nielsen J, Villadsen J (2000) Optimization of ethanol production in Saccharomyces cerevisiae by metabolic engineering of the ammonium assimilation. Metab Eng 2:69–77. https://doi.org/10.1006/mben.1999.0140
Nygård Y, Mojzita D, Toivari M, Penttilä M, Wiebe MG, Ruohonen L (2014) The diverse role of Pdr12 in resistance to weak organic acids. Yeast 31:219–232. https://doi.org/10.1002/yea.3011
Oughtred R, Chatr-Aryamontri A, Breitkreutz BJ, Chang CS, Rust JM, Theesfeld CL, Heinicke S, Breitkreutz A, Chen D, Hirschman J, Kolas N, Livstone MS, Nixon J, O’Donnell L, Ramage L, Winter A, Reguly T, Sellam A, Stark C, Boucher L, Dolinski K, Tyers M (2016) Biogrid: a resource for studying biological interactions in yeast. Cold Spring Harb Protoc 2016:29–34. https://doi.org/10.1101/pdb.top080754
Overkamp KM, Kötter P, van der Hoek R, Schoondermark-Stolk S, Luttik MAH, van Dijken JP, Pronk JT (2002) Functional analysis of structural genes for NAD(+)-dependent formate dehydrogenase in Saccharomyces cerevisiae. Yeast 19:509–520. https://doi.org/10.1002/yea.856
Palmqvist E, Grage H, Meinander NQ, Hahn-Hägerdal B (1999) Main and interaction effects of acetic acid, furfural, and p-hydroxybenzoic acid on growth and ethanol productivity of yeasts. Biotechnol Bioeng 63:46–55. https://doi.org/10.1002/(sici)1097-0290(19990405)63:1<46::aid-bit5>3.0.co;2-j
Palmqvist E, Hahn-Hägerdal B (2000) Fermentation of lignocellulosic hydrolysates. II: inhibitors and mechanisms of inhibition. Bioresour Technol 74:25–33. https://doi.org/10.1016/S0960-8524(99)00161-3
Pampulha ME, Loureiro-Dias MC (1989) Combined effect of acetic acid, pH and ethanol on intracellular pH of fermenting yeast. Appl Microbiol Biotechnol 31:547–550. https://doi.org/10.1007/BF00270792
Parawira W, Tekere M (2011) Biotechnological strategies to overcome inhibitors in lignocellulose hydrolysates for ethanol production: review. Crit Rev Biotechnol 31:20–31. https://doi.org/10.3109/07388551003757816
Piper MD, Hong S-P, Ball GE, Dawes IW (2000) Regulation of the Balance of One-carbon Metabolism inSaccharomyces cerevisiae. J Biol Chem 275:30987–30995. https://doi.org/10.1074/jbc.M004248200
Ravindran R, Jaiswal AK (2016) A comprehensive review on pre-treatment strategy for lignocellulosic food industry waste: challenges and opportunities. Bioresour Technol 199:92–102. https://doi.org/10.1016/j.biortech.2015.07.106
Russell JB (1992) Another explanation for the toxicity of fermentation acids at low pH: anion accumulation versus uncoupling. J Appl Bacteriol 73:363–370. https://doi.org/10.1111/j.1365-2672.1992.tb04990.x
Sanda T, Hasunuma T, Matsuda F, Kondo A (2011) Repeated-batch fermentation of lignocellulosic hydrolysate to ethanol using a hybrid Saccharomyces cerevisiae strain metabolically engineered for tolerance to acetic and formic acids. Bioresour Technol 102:7917–7924. https://doi.org/10.1016/j.biortech.2011.06.028
Schuller C, Mamnun YM, Mollapour M, Krapf G, Schuster M, Bauer BE, Piper PW, Kuchler K (2004) Global phenotypic analysis and transcriptional profiling defines the weak acid stress response regulon in Saccharomyces cerevisiae. Mol Biol Cell 15:706–720
Shashkova S, Welkenhuysen N, Hohmann S (2015) Molecular communication: crosstalk between the Snf1 and other signaling pathways. FEMS Yeast Res. https://doi.org/10.1093/femsyr/fov026
Sutherland FC, Lages F, Lucas C, Luyten K, Albertyn J, Hohmann S, Prior BA, Kilian SG (1997) Characteristics of Fps1-dependent and -independent glycerol transport in Saccharomyces cerevisiae. J Bacteriol 179:7790–7795. https://doi.org/10.1128/jb.179.24.7790-7795.1997
Taherzadeh MJ, Eklund R, Gustafsson L, Niklasson C, Lidén G (1997) Characterization and fermentation of dilute-acid hydrolyzates from wood. Ind Eng Chem Res 36:4659–4665. https://doi.org/10.1021/ie9700831
Tanaka K, Ishii Y, Ogawa J, Shima J (2012) Enhancement of acetic acid tolerance in Saccharomyces cerevisiae by overexpression of the HAA1 gene, encoding a transcriptional activator. Appl Environ Microbiol 78:8161–8163. https://doi.org/10.1128/AEM.02356-12
Turcotte B, Liang XB, Robert F, Soontorngun N (2010) Transcriptional regulation of nonfermentable carbon utilization in budding yeast. FEMS Yeast Res 10:2–13. https://doi.org/10.1111/j.1567-1364.2009.00555.x
Vemuri GN, Eiteman MA, McEwen JE, Olsson L, Nielsen J (2007) Increasing NADH oxidation reduces overflow metabolism in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 104:2402–2407. https://doi.org/10.1073/pnas.0607469104
Viegas CA, Almeida PF, Cavaco M, Sa-Correia I (1998) The H + -ATPase in the plasma membrane of Saccharomyces cerevisiae is activated during growth latency in octanoic acid-supplemented medium accompanying the decrease in intracellular pH and cell viability. Appl Envir Microbiol 64:779–783
Viegas CA, Sá-Correia I (1991) Activation of plasma membrane ATPase of Saccharomyces cerevisiae by octanoic acid. J Gen Microbiol 137:645–651. https://doi.org/10.1099/00221287-137-3-645
Wei N, Quarterman J, Kim SR, Cate JHD, Jin Y-S (2013) Enhanced biofuel production through coupled acetic acid and xylose consumption by engineered yeast. Nat Commun 4:2580. https://doi.org/10.1038/ncomms3580
Yao Y, Tsuchiyama S, Yang C, Bulteau AL, He C, Robison B, Tsuchiya M, Miller D, Briones V, Tar K, Potrero A, Friguet B, Kennedy BK, Schmidt M (2015) Proteasomes, Sir2, and Hxk2 form an interconnected aging network that impinges on the AMPK/Snf1-regulated transcriptional repressor Mig1. PLoS Genet. https://doi.org/10.1371/journal.pgen.1004968
Zhang JG, Liu XY, He XP, Guo XN, Lu Y, Zhang BR (2011) Improvement of acetic acid tolerance and fermentation performance of Saccharomyces cerevisiae by disruption of the FPS1 aquaglyceroporin gene. Biotechnol Lett 33:277–284. https://doi.org/10.1007/s10529-010-0433-3
Zhang M, Galdieri L, Vancura A (2013) The yeast AMPK homolog SNF1 regulates acetyl coenzyme A homeostasis and histone acetylation. Mol Cell Biol 33:4701–4717. https://doi.org/10.1128/MCB.00198-13
Zheng DQ, Wu XC, Wang PM, Chi XQ, Tao XL, Li P, Jiang XH, Zhao YH (2011) Drug resistance marker-aided genome shuffling to improve acetic acid tolerance in Saccharomyces cerevisiae. J Ind Microbiol Biotechnol 38:415–422. https://doi.org/10.1007/s10295-010-0784-8
Acknowledgements
Authors would like to thank Prof. Vince Martin for S. cerevisiae CEN.PK 113-7D strain. Authors would like to acknowledge funding and postdoctoral scholarship (VEBH) from Agricultural Bioproducts Innovation Program, NSERC Bioconversion Network and BioFuelNet.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Balderas-Hernández, V.E., Correia, K. & Mahadevan, R. Inactivation of the transcription factor mig1 (YGL035C) in Saccharomyces cerevisiae improves tolerance towards monocarboxylic weak acids: acetic, formic and levulinic acid. J Ind Microbiol Biotechnol 45, 735–751 (2018). https://doi.org/10.1007/s10295-018-2053-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-018-2053-1