Abstract
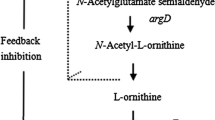
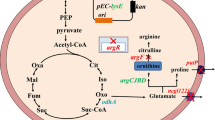
l-Ornithine is a non-protein amino acid with extensive applications in the food and pharmaceutical industries. In this study, we performed metabolic pathway engineering of an l-arginine hyper-producing strain of Corynebacterium crenatum for L-ornithine production. First, we amplified the L-ornithine biosynthetic pathway flux by blocking the competing branch of the pathway. To enhance L-ornithine synthesis, we performed site-directed mutagenesis of the ornithine-binding sites to solve the problem of l-ornithine feedback inhibition for ornithine acetyltransferase. Alternatively, the genes argA from Escherichia coli and argE from Serratia marcescens, encoding the enzymes N-acetyl glutamate synthase and N-acetyl-l-ornithine deacetylase, respectively, were introduced into Corynebacterium crenatum to mimic the linear pathway of L-ornithine biosynthesis. Fermentation of the resulting strain in a 5-L bioreactor allowed a dramatically increased production of L-ornithine, 40.4 g/L, with an overall productivity of 0.673 g/L/h over 60 h. This demonstrates that an increased level of transacetylation is beneficial for L-ornithine biosynthesis.





Similar content being viewed by others
References
Amund OO, Mackinnon G, Higgins IJ (1983) Increased l-ornithine production by an arg mutant of Acinetobacter lwoffi. Eur J Appl Microbiol Biotechnol 17:252–253
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201. https://doi.org/10.1093/bioinformatics/bti770
Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258. https://doi.org/10.1093/nar/gku340
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Casteele MVD, Demarez M, Legrain C, Glansdorff N, Piérard A (1990) Pathways of arginine biosynthesis in extreme thermophilic archeo- and eubacteria. J Gen Microbiol 136:1177–1183
Choi DK, Ryu WS, Choi CY, Park YH (1996) Production of l-ornithine by arginine auxotrophic mutants of Brevibacterium ketoglutamicum in dual substrate-limited continuous culture. J Ferment Bioeng 81:216–219
Delano WL (2002) The PyMOL Molecular Graphics System. https://pymol.org
Dyakonov T, Muir A, Nasri H, Toops D, Fatmi A (2013) Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat Methods 10:659–664
Errey JC, Blanchard JS (2005) Functional characterization of a novel argA from Mycobacterium tuberculosis. J Bacteriol 187:3039–3044. https://doi.org/10.1128/JB.187.9.3039-3044.2005
Han PS, Fishel RS, Efron DT, Williams JZ, Fishel MH, Barbul A (2002) Effect of supplemental ornithine on wound healing. J Surg Res 106:299
Hermann SG (2006) Tricine-SDS-PAGE. Nat Protoc 1:16–22
Hwang JH, Hwang GH, Cho JY (2008) Effect of increased glutamate availability on l-ornithine production in Corynebacterium glutamicum. J Microbiol Biotechnol 18:704
Javidmajd F, Blanchard JS (2000) Mechanistic analysis of the argE-encoded N-acetylornithine deacetylase. Biochemistry 39:1285
Jiang LY, Chen SG, Zhang YY, Liu JZ (2013) Metabolic evolution of Corynebacterium glutamicum for increased production of l-ornithine. BMC Biotechnol 13:47–47
Jiang LY, Zhang YY, Li Z, Liu JZ (2013) Metabolic engineering of Corynebacterium glutamicum for increasing the production of l-ornithine by increasing NADPH availability. J Ind Microbiol Biotechnol 40:1143–1151
Kim SY, Lee J, Lee SY (2015) Metabolic engineering of Corynebacterium glutamicum for the production of l-ornithine. Biotechnol Bioeng 112:416–421. https://doi.org/10.1002/bit.25440
Kinoshita S, Nakayama K, Udaka S (2006) The fermentative production of l-ornithine: preliminary report. J Gen Appl Microbiol 3:276–277
Lee JH, Wendisch VF (2017) Production of amino acids - Genetic and metabolic engineering approaches. Bioresour Technol 245:1575
Lee YJ, Cho JY (2006) Genetic manipulation of a primary metabolic pathway for l-ornithine production in Escherichia coli. Biotechnol Lett 28:1849–1856. https://doi.org/10.1007/s10529-006-9163-y
Leisinger T, Haas D (1975) N-Acetylglutamate synthase of Escherichia coli regulation of synthesis and activity by arginine. J Biol Chem 250:1690–1693
Liu Y, Heeswijck RV, Høj P, Hoogenraad N (1995) Purification and characterization of ornithine acetyltransferase from Saccharomyces cerevisiae. Eur J Biochem 228:291–296
Man Z, Xu M, Rao Z, Guo J, Yang T, Zhang X, Xu Z (2016) Systems pathway engineering of Corynebacterium crenatum for improved l-arginine production. Sci Rep 6:28629. https://doi.org/10.1038/srep28629
Marc F, Weigel P, Legrain C, Almeras Y, Santrot M, Glansdorff N, Sakanyan V (2010) Characterization and kinetic mechanism of mono- and bifunctional ornithine acetyltransferases from thermophilic microorganisms. Febs J 267:5217–5226
Zhao G, Haskins N, Jin Z, N MA, Tuchman M, Shi D (2013) Structure of N-acetyl-l-glutamate synthase/kinase from Maricaulis maris with the allosteric inhibitor l-arginine bound. Biochem Biophys Res Commun 437:585–590. https://doi.org/10.1016/j.bbrc.2013.07.003
Mcgregor WC, Gillner DM, Swierczek SI, Liu D, Holz RC (2013) Identification of a histidine metal ligand in the argE-Encoded N-acetyl-l-ornithine deacetylase from Escherichia coli. Springerplus 2:482
Morris GM, Ruth H, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) Software news and updates AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791
Mutalik VK, Guimaraes JC, Cambray G, Lam C, Christoffersen MJ, Mai QA, Tran AB, Paull M, Keasling JD, Arkin AP, Endy D (2013) Precise and reliable gene expression via standard transcription and translation initiation elements. Nat Methods 10:354–360. https://doi.org/10.1038/nmeth.2404
Nowroozi FF, Baidoo EE, Ermakov S, Redding-Johanson AM, Batth TS, Petzold CJ, Keasling JD (2014) Metabolic pathway optimization using ribosome binding site variants and combinatorial gene assembly. Appl Microbiol Biotechnol 98:1567–1581
Qian ZG, Xia XX, Sangyup L (2009) Metabolic engineering of Escherichia coli for the production of putrescine: a four carbon diamine. Biotechnol Bioeng 104:651–662
Qin J, Zhou YJ, Krivoruchko A, Huang M, Liu L, Khoomrung S, Siewers V, Jiang B, Nielsen J (2015) Modular pathway rewiring of Saccharomyces cerevisiae enables high-level production of l-ornithine. Nat Commun 6:8224. https://doi.org/10.1038/ncomms9224
Rajagopal BS, Deponte J, Tuchman M, Malamy MH (1998) Use of inducible feedback-resistant N-acetylglutamate synthetase (argA) genes for enhanced arginine biosynthesis by genetically engineered Escherichia coli K-12 Strains. Appl Environ Microbiol 64:1805
Sakanyan V, Petrosyan P, Lecocq M, Boyen A, Legrain C, Demarez M, Hallet JN, Glansdorff N (1996) Genes and enzymes of the acetyl cycle of arginine biosynthesis in Corynebacterium glutamicum: enzyme evolution in the early steps of the arginine pathway. Microbiology 142(Pt 1):99–108. https://doi.org/10.1099/13500872-142-1-99
Salis HM, Mirsky EA, Voigt CA (2009) Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol 27:946–950. https://doi.org/10.1038/nbt.1568
Salvatore F, Cimino F, d’Ayello-Caracciolo M, Cittadini D (1964) Mechanism of the protection by l-ornithine-l-aspartate mixture and by l-arginine in ammonia intoxication. Arch Biochem Biophys 107:499–503. https://doi.org/10.1016/0003-9861(64)90307-8
Sambrook J, Fritsch EF, Maniatis T (2001) Molecular cloning. A laboratory manual. Anal Biochem 186:182–183
Sancho E (2009) Mechanism of arginine regulation of acetylglutamate synthase, the first enzyme of arginine synthesis. Febs Lett 583:202–206
Sankaranarayanan R, Cherney MM, Garen C, Garen G, Niu C, Yuan M, James MN (2010) The molecular structure of ornithine acetyltransferase from Mycobacterium tuberculosis bound to ornithine, a competitive inhibitor. J Mol Biol 397:979–990. https://doi.org/10.1016/j.jmb.2010.02.018
Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A (1994) Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69
Tauch A, Kirchner O, Löffler B, Götker S, Pühler A, Kalinowski J (2002) Efficient electrotransformation of Corynebacterium diphtheriae with a mini-replicon derived from the Corynebacterium glutamicum plasmid pGA1. Curr Microbiol 45:362–367
Velasco AM, Leguina JI, Lazcano A (2002) Molecular evolution of the lysine biosynthetic pathways. J Mol Evol 55:445–449. https://doi.org/10.1007/s00239-002-2340-2
Xu H, Dou W, Xu H, Zhang X, Rao Z, Shi Z, Xu Z (2009) A two-stage oxygen supply strategy for enhanced l-arginine production by Corynebacterium crenatum based on metabolic fluxes analysis. Biochem Eng J 43:41–51. https://doi.org/10.1016/j.bej.2008.08.007
Xu M, Rao Z, Dou W, Xu Z (2013) The role of ARGR repressor regulation on l-arginine production in Corynebacterium crenatum. Appl Biochem Biotechnol 170:587–597. https://doi.org/10.1007/s12010-013-0212-4
Zhang B, Yu M, Zhou Y, Li Y, Ye BC (2017) Systematic pathway engineering of Corynebacterium glutamicum S9114 for l-ornithine production. Microb Cell Fact 16:158. https://doi.org/10.1186/s12934-017-0776-8
Zhang B, Zhou N, Liu YM, Liu C, Lou CB, Jiang CY, Liu SJ (2015) Ribosome binding site libraries and pathway modules for shikimic acid synthesis with Corynebacterium glutamicum. Microb Cell Fact 14:71. https://doi.org/10.1186/s12934-015-0254-0
Zhang J, Xu M, Ge X, Zhang X, Yang T, Xu Z, Rao Z (2016) Reengineering of the feedback-inhibition enzyme N-acetyl-l-glutamate kinase to enhance l-arginine production in Corynebacterium crenatum. J Ind Microbiol Biotechnol 44:271–283. https://doi.org/10.1007/s10295-016-1885-9
Zhao Q, Luo Y, Dou W, Zhang X, Zhang X, Zhang W, Xu M, Geng Y, Rao Z, Xu Z (2016) Controlling the transcription levels of argGH redistributed l-arginine metabolic flux in N-acetylglutamate kinase and ArgR-deregulated Corynebacterium crenatum. J Ind Microbiol Biotechnol 43:55–66
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31770058, 31570085), the Jiangsu Province Science Fund for Distinguished Young Scholars (BK20150002), the Research Project of the Chinese Ministry of Education (113033A), the Fundamental Research Funds for the Central Universities (JUSRP51708A), the national first-class discipline program of Light Industry Technology and Engineering (LITE2018-06) and the 111 Project (111-2-06).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shu, Q., Xu, M., Li, J. et al. Improved l-ornithine production in Corynebacterium crenatum by introducing an artificial linear transacetylation pathway. J Ind Microbiol Biotechnol 45, 393–404 (2018). https://doi.org/10.1007/s10295-018-2037-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-018-2037-1