Abstract
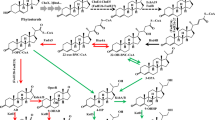
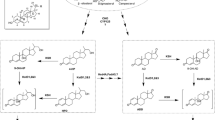
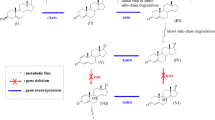
Mycobacterium neoaurum ST-095 and its mutant M. neoaurum JC-12, capable of transforming phytosterol to androst-1,4-diene-3,17-dione (ADD) and androst-4-ene-3,17-dione (AD), produce very different molar ratios of ADD/AD. The distinct differences were related to the enzyme activity of 3-ketosteroid-Δ1-dehydrogenase (KSDD), which catalyzes the C1,2 dehydrogenation of AD to ADD specifically. In this study, by analyzing the primary structure of KSDDI (from M. neoaurum ST-095) and KSDDII (from M. neoaurum JC-12), we found the only difference between KSDDI and KSDDII was the mutation of Val366 to Ser366. This mutation directly affected KSDD enzyme activity, and this result was confirmed by heterologous expression of these two enzymes in Bacillus subtilis. Assay of the purified recombinant enzymes showed that KSDDII has a higher C1,2 dehydrogenation activity than KSDDI. The functional difference between KSDDI and KSDDII in phytosterol biotransformation was revealed by gene disruption and complementation. Phytosterol transformation results demonstrated that ksdd I and ksdd II gene disrupted strains showed similar ADD/AD molar ratios, while the ADD/AD molar ratios of the ksdd I and ksdd II complemented strains were restored to their original levels. These results proved that the different ADD/AD molar ratios of these two M. neoaurum strains were due to the differences in KSDD. Finally, KSDD structure analysis revealed that the Val366Ser mutation could possibly play an important role in stabilizing the active center and enhancing the interaction of AD and KSDD. This study provides a reliable theoretical basis for understanding the structure and catalytic mechanism of the Mycobacteria KSDD enzyme.







Similar content being viewed by others
Abbreviations
- AD:
-
Androst-4-ene-3,17-dione
- ADD:
-
Androst-1,4-diene-3,17-dione
- KSDD:
-
3-Ketosteroid-Δ1-dehydrogenase
- FAD:
-
Flavin adenine dinucleotide
- Me-β-CD:
-
Methyl-β-cyclodextrin
- PMS:
-
Phenazine methosulphate
- DCPIP:
-
2,6-Dichlorophenolindophenol
References
Anagnostopoulos C, Spizizen J (1961) Requirements for transformation in Bacillus subtilis. J Bacteriol 81:741
Bragin EY, Shtratnikova VY, Dovbnya D, Schelkunov M, Pekov YA, Malakho S, Egorova O, Ivashina T, Sokolov S, Ashapkin V (2013) Comparative analysis of genes encoding key steroid core oxidation enzymes in fast-growing Mycobacterium spp. strains. J Steroid Biochem Mol Biol 138:41–53
Choi K-P, Molnar I, Yamashita M, Murooka Y (1995) Purification and characterization of the 3-ketosteroid-Δ1-dehydrogenase of Arthrobacter simplex produced in Streptomyces lividans. J Biochem 117:1043–1049
Donova MV, Egorova OV (2012) Microbial steroid transformations: current state and prospects. Appl Microbiol Biotechnol 94:1423–1447. doi:10.1007/s00253-012-4078-0
Fernandes P, Cruz A, Angelova B, Pinheiro HM, Cabral JMS (2003) Microbial conversion of steroid compounds: recent developments. Enzyme Microb Technol 32:688–705. doi:10.1016/S0141-0229(03)00029-2
Fujii C, Morii S, Kadode M, Sawamoto S, Iwami M, Itagaki E (1999) Essential tyrosine residues in 3-Ketosteroid-Δ’-Dehydrogenase from Rhodococcus rhodochrous. J Biochem 126:662–667
Gordhan BG, Parish T (2001) Gene replacement using pretreated DNA. In: Parish T, Stoker NG (eds) Mycobacterium tuberculosis protocols, vol 54. Methods in Molecular Medicine. Humana Press Inc., New Jersey, pp 77–92. doi:10.1385/1-59259-147-7:077
Li Y, Lu F, Sun T, Du L (2007) Expression of ksdD gene encoding 3-ketosteroid-Δ1-dehydrogenase from Arthrobacter simplex in Bacillus subtilis. Lett Appl Microbiol 44:563–568
Liu C, Zhang X, Z-m Rao, Shao M-l WuD, Z-h Xu, Li H (2015) Mutation breeding of high 4-androstene-3, 17-dione-producing Mycobacterium neoaurum ZADF-4 by atmospheric and room temperature plasma treatment. J Zhejiang Univ Sci B 16:286–295
Long S, Zhang X, Rao Z, Chen K, Xu M, Yang T, Yang S (2016) Amino acid residues adjacent to the catalytic cavity of tetramer l-asparaginase II contribute significantly to its catalytic efficiency and thermostability. Enzyme Microb Technol 82:15–22. doi:10.1016/j.enzmictec.2015.08.009
Malaviya A, Gomes J (2008) Androstenedione production by biotransformation of phytosterols. Bioresour Technol 99:6725–6737. doi:10.1016/j.biortech.2008.01.039
Marsheck WJ, Kraychy S, Muir RD (1972) Microbial degradation of sterols. Appl Microbiol 23:72–77
Martin CK (1977) Microbial cleavage of sterol side chains. Adv Appl Microbiol 22:29–58
Matsushita H, Itagaki E (1992) Essential histidine residue in 3-ketosteroid-Δ1-dehydrogenase. J Biochem 111:594–599
Molnar I, Choi KP, Yamashita M, Murooka Y (1995) Molecular cloning, expression in Streptomyces livdans, and analysis of a gene cluster from Arthrobacter simplex encoding 3-ketosteroid-Δ1-dehydrogenase, 3-ketosteroid-Δ5-isomerase and a hypothetical regulatory protein. Mol Microbiol 15:895–905
Morii S, Fujii C, Miyoshi T, Iwami M, Itagaki E (1998) 3-Ketosteroid-Δ1-dehydrogenase of Rhodococcus rhodochrous: sequencing of the genomic DNA and hyperexpression, purification, and characterization of the recombinant enzyme. J Biochem 124:1026–1032
Murray HC, Peterson DH (1952) Oxygenation of steroids by Mucorales fungi. Google Patents
Plesiat P, Grandguillot M, Harayama S, Vragar S, Michel-Briand Y (1991) Cloning, sequencing, and expression of the Pseudomonas testosteroni gene encoding 3-oxosteroid delta 1-dehydrogenase. J Bacteriol 173:7219–7227
Rohman A, van Oosterwijk N, Thunnissen A-MW, Dijkstra BW (2013) Crystal structure and site-directed mutagenesis of 3-ketosteroid Δ1-dehydrogenase from Rhodococcus erythropolis SQ1 explain its catalytic mechanism. J Biol Chem 288:35559–35568
Shao M, Rao Z, Zhang X, Xu M, Yang T, Li H, Xu Z, Yang S (2015) Bioconversion of cholesterol to 4-cholesten-3-one by recombinant Bacillus subtilis expressing choM gene encoding cholesterol oxidase from Mycobacterium neoaurum JC-12. J Chem Technol Biotechnol 90:1811–1820. doi:10.1002/jctb.4491
Shao M, Zhang X, Rao Z, Xu M, Yang T, Li H, Xu Z (2015) Enhanced production of Androst-1, 4-Diene-3, 17-Dione by Mycobacterium neoaurum JC-12 using three-stage fermentation strategy. PLoS One 10:e0137658
Shao M, Zhang X, Rao Z, Xu M, Yang T, Li H, Xu Z, Yang S-T (2015) Efficient testosterone production by engineered Pichia pastoris co-expressing human 17β-hydroxysteroid dehydrogenase type 3 and Saccharomyces cerevisiae glucose 6-phosphate dehydrogenase with NADPH regeneration. Green Chem. doi:10.1039/C5GC02353J
Shen YB, Wang M, Li HN, Wang YB, Luo JM (2012) Influence of hydroxypropyl-β-cyclodextrin on phytosterol biotransformation by different strains of Mycobacterium neoaurum. J Ind Microbiol Biotechnol 39:1253–1259
Van der Geize R, Hessels G, Van Gerwen R, Vrijbloed J, Van der Meijden P, Dijkhuizen L (2000) Targeted disruption of the kstD gene encoding a 3-Ketosteroid Δ1-Dehydrogenase isoenzyme of rhodococcus erythropolis strain SQ1. Appl Environ Microbiol 66:2029–2036
van Oosterwijk N, Knol J, Dijkhuizen L, van der Geize R, Dijkstra BW (2012) Structure and catalytic mechanism of 3-ketosteroid-Δ4-(5α)-dehydrogenase from Rhodococcus jostii RHA1 genome. J Biol Chem 287:30975–30983
Wang Z, Zhao F, Chen D, Li D (2006) Biotransformation of phytosterol to produce androsta-diene-dione by resting cells of Mycobacterium in cloud point system. Process Biochem 41:557–561
Wang ZF, Huang YL, Rathman JF, Yang ST (2002) Lecithin-enhanced biotransformation of cholesterol to androsta-1, 4-diene-3, 17-dione and androsta-4-ene-3, 17-dione. J Chem Technol Biotechnol 77:1349–1357
Wei W, Wang FQ, Fan SY, Wei DZ (2010) Inactivation and augmentation of the primary 3-ketosteroid-Δ1-dehydrogenase in Mycobacterium neoaurum NwIB-01: biotransformation of soybean phytosterols to 4-androstene-3,17-dione or 1,4-androstadiene-3,17-dione. Appl Environ Microbiol 76:4578–4582. doi:10.1128/aem.00448-10
Xie RL, Shen YB, Qin N, Wang YB, Su LQ, Wang M (2015) Genetic differences in ksdD influence on the ADD/AD ratio of Mycobacterium neoaurum. J Ind Microbiol Biotechnol:1–7
Xu XX, Jin FL, Yu XQ, Ji SX, Wang J, Cheng HX, Wang C, Zhang WQ (2007) Expression and purification of a recombinant antibacterial peptide, cecropin, from Escherichia coli. Protein Expr Purif 53:293–301
Xu Y-P, Qin J, Sun S-M, Liu T-T, Zhang X-L, Qian S-S, Zhu H-L (2014) Synthesis, crystal structures, molecular docking and urease inhibitory activity of nickel (II) complexes with 3-pyridinyl-4-amino-5-mercapto-1, 2, 4-triazole. Inorganica Chim Acta 423:469–476
Yao K, Xu L-Q, Wang F-Q, Wei D-Z (2014) Characterization and engineering of 3-ketosteroid-△1-dehydrogenase and 3-ketosteroid-9α-hydroxylase in Mycobacterium neoaurum ATCC 25795 to produce 9α-hydroxy-4-androstene-3,17-dione through the catabolism of sterols. Metab Eng 24:181–191. doi:10.1016/j.ymben.2014.05.005
Zhang WQ, Shao ML, Rao ZM, Xu MJ, Zhang X, Yang TW, Li H, Xu ZH (2013) Bioconversion of 4-androstene-3,17-dione to androst-1,4-diene-3,17-dione by recombinant Bacillus subtilis expressing ksdd gene encoding 3-ketosteroid-Δ1-dehydrogenase from Mycobacterium neoaurum JC-12. J Steroid Biochem Mol Biol 135:36–42. doi:10.1016/j.jsbmb.2012.12.016
Zhong C, Rao Z, Xia H, Xu Z, Fang H, Zhuge J (2009) Mutation breeding of Mycobacterium sp. for transformation of phytosterol into androst-1,4-diene-3,17-dione. Chem Bioeng 7:43–46 (in Chinese)
Acknowledgments
We sincerely appreciate Professor W. R. Jacobs, Jr. (Howard Hughes Medical Institute, USA) for providing plasmids pMV261 and pMV306, and Professor T. Parish (Department of Infectious & Tropical Diseases, United Kingdom) for providing plasmids of p2NIL and pGOAL19. This work was supported by the National Basic Research Program of China (973 Program) (2012CB725202), the High-Tech Research and Development Programs of China (2011AA02A211, SS2015AA021004), the National Natural Science Foundation of China (31570085), Jiangsu Province Science Fund for Distinguished Young Scholars (BK20150002), the Fundamental Research Funds for the Central Universities (JUSRP51306A), the 111 Project (111-2-06) and a Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shao, M., Zhang, X., Rao, Z. et al. A mutant form of 3-ketosteroid-Δ1-dehydrogenase gives altered androst-1,4-diene-3, 17-dione/androst-4-ene-3,17-dione molar ratios in steroid biotransformations by Mycobacterium neoaurum ST-095. J Ind Microbiol Biotechnol 43, 691–701 (2016). https://doi.org/10.1007/s10295-016-1743-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-016-1743-9