Abstract
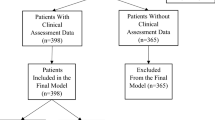
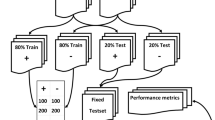
A significant volume of medical data remains unstructured. Natural language processing (NLP) and machine learning (ML) techniques have shown to successfully extract insights from radiology reports. However, the codependent effects of NLP and ML in this context have not been well-studied. Between April 1, 2015 and November 1, 2016, 9418 cross-sectional abdomen/pelvis CT and MR examinations containing our internal structured reporting element for cancer were separated into four categories: Progression, Stable Disease, Improvement, or No Cancer. We combined each of three NLP techniques with five ML algorithms to predict the assigned label using the unstructured report text and compared the performance of each combination. The three NLP algorithms included term frequency-inverse document frequency (TF-IDF), term frequency weighting (TF), and 16-bit feature hashing. The ML algorithms included logistic regression (LR), random decision forest (RDF), one-vs-all support vector machine (SVM), one-vs-all Bayes point machine (BPM), and fully connected neural network (NN). The best-performing NLP model consisted of tokenized unigrams and bigrams with TF-IDF. Increasing N-gram length yielded little to no added benefit for most ML algorithms. With all parameters optimized, SVM had the best performance on the test dataset, with 90.6 average accuracy and F score of 0.813. The interplay between ML and NLP algorithms and their effect on interpretation accuracy is complex. The best accuracy is achieved when both algorithms are optimized concurrently.


Similar content being viewed by others
References
Schwartz LH, Panicek DM, Berk AR, Li Y, Hricak H: Improving communication of diagnostic radiology findings through structured reporting. Radiology. 260(1):174–181, 2011
Lakhani P, Kim W, Langlotz CP: Automated extraction of critical test values and communications from unstructured radiology reports: an analysis of 9.3 million reports from 1990 to 2011. Radiology. 265(3):809–818, 2012
Lakhani P, Kim W, Langlotz CP: Automated detection of critical results in radiology reports. J Digit Imaging. 25(1):30–36, 2012
Cai T, Giannopoulos AA, Yu S, Kelil T, Ripley B, Kumamaru KK et al.: Natural language processing technologies in radiology research and clinical applications. Radiogr Rev Publ Radiol Soc N Am Inc. 36(1):176–191, 2016
Yim W-W, Yetisgen M, Harris WP, Kwan SW: Natural language processing in oncology: a review. JAMA Oncol. 2(6):797–804, 2016
Kocbek S, Cavedon L, Martinez D, Bain C, Mac Manus C, Haffari G et al.: Text mining electronic hospital records to automatically classify admissions against disease: measuring the impact of linking data sources. J Biomed Inform., 2016
Rajaraman A, Ullman JD: Mining of massive datasets [Internet]. Cambridge: Cambridge University Press, 2011, [cited 2017 May 24]. Available from: http://ebooks.cambridge.org/ref/id/CBO9781139058452
Porter MF: An algorithm for suffix stripping. Program. 14(3):130–137, 1980
Wu HC, Luk RWP, Wong KF, Kwok KL: Interpreting TF-IDF term weights as making relevance decisions. ACM Trans Inf Syst. 26(3):1–37, 2008
Weinberger K, Dasgupta A, Langford J, Smola A, Attenberg J: Feature hashing for large scale multitask learning. In ACM Press; 2009 [cited 2017 May 24]. p. 1–8. Available from: http://portal.acm.org/citation.cfm?doid=1553374.1553516
Hassanpour S, Langlotz CP. Unsupervised Topic Modeling in a Large Free Text Radiology Report Repository. J Digit Imaging. 29(1):59–62, 2016.
Hassanpour S, Langlotz CP. Information extraction from multi-institutional radiology reports. Artif Intell Med. 66:29–39, 2016.
Horng S, Sontag DA, Halpern Y, Jernite Y, Shapiro NI, Nathanson LA: Creating an automated trigger for sepsis clinical decision support at emergency department triage using machine learning. PloS One. 12(4):e0174708, 2017
Morid MA, Fiszman M, Raja K, Jonnalagadda SR, Del Fiol G: Classification of clinically useful sentences in clinical evidence resources. J Biomed Inform. 60:14–22, 2016
Polak S, Mendyk A: Artificial neural networks as an engine of Internet based hypertension prediction tool. Stud Health Technol Inform. 103:61–69, 2004
Tong W, Xie Q, Hong H, Shi L, Fang H, Perkins R et al.: Using decision forest to classify prostate cancer samples on the basis of SELDI-TOF MS data: assessing chance correlation and prediction confidence. Environ Health Perspect. 112(16):1622–1627, 2004
Wang Z, He Y, Jiang M: A Comparison among Three Neural Networks for Text Classification. In IEEE; 2006 [cited 2017 May 29]. Available from: http://ieeexplore.ieee.org/document/4129218/
Zafar HM, Chadalavada SC, Kahn CE, Cook TS, Sloan CE, Lalevic D et al.: Code abdomen: an assessment coding scheme for abdominal imaging findings possibly representing cancer. J Am Coll Radiol JACR. 12(9):947–950, 2015
Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L et al.: New guidelines to evaluate the response to treatment in solid tumors. J Natl Cancer Inst. 92(3):205–216, 2000
Bird S, Klein E, Loper E: Natural language processing with Python, 1st edition. Beijing: O’Reilly, 2009, 479 p
Lipton ZC, Elkan C, Naryanaswamy B: Optimal thresholding of classifiers to maximize F1 measure. Mach Learn Knowl Discov Databases Eur Conf ECML PKDD Proc ECML PKDD Conf. 8725:225–239, 2014
Bennasar M, Hicks Y, Setchi R: Feature selection using joint mutual information maximisation. Expert Syst Appl. 42(22):8520–8532, 2015
Hripcsak G, Rothschild AS: Agreement, the f-measure, and reliability in information retrieval. J Am Med Inform Assoc JAMIA. 12(3):296–298, 2005
Pennington J, Socher R, Manning C. Glove: global vectors for word representation. In association for computational linguistics; 2014 [cited 2017 Sep 2]. p. 1532–43. Available from: http://aclweb.org/anthology/D14-1162
Mikolov T, Chen K, Corrado G, Dean J. Efficient Estimation of Word Representations in Vector Space. ICLR Workshop. 2013 Jan 16;
Zhang W, Yoshida T, Tang X. TFIDF, LSI and multi-word in information retrieval and text categorization. In IEEE; 2008 [cited 2017 May 29]. p. 108–13. Available from: http://ieeexplore.ieee.org/document/4811259/
Dietrich R, Opper M, Sompolinsky H: Statistical mechanics of support vector networks. Phys Rev Lett. 82(14):2975–2978, 1999
Joachims T: Text categorization with support vector machines: learning with many relevant features. In: Nédellec C, Rouveirol C Eds. Machine learning: ECML-98 [Internet]. Berlin: Springer Berlin Heidelberg, 1998, pp. 137–142 [cited 2017 May 29] Available from: http://link.springer.com/10.1007/BFb0026683
Liu X, Song M, Tao D, Liu Z, Zhang L, Chen C et al.: Random forest construction with robust semisupervised node splitting. IEEE Trans Image Process Publ IEEE Signal Process Soc. 24(1):471–483, 2015
Wang J, Zhang J, An Y, Lin H, Yang Z, Zhang Y et al.: Biomedical event trigger detection by dependency-based word embedding. BMC Med Genomics 9 Suppl 2:45, 2016
Wei W, Marmor R, Singh S, Wang S, Demner-Fushman D, Kuo T-T et al.: Finding related publications: extending the set of terms used to assess article similarity. AMIA Jt Summits Transl Sci Proc AMIA Jt Summits Transl Sci. 2016:225–234, 2016
Funding
This study received no funding support from a grant agency.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Po-Hao Chen is a co-founder of Alphametric Health LLC. Maya Galperin-Aizenberg, Hanna Zafar, and Tessa S. Cook declare that they have no conflicts of interest.
Informed Consent
For this type of study formal consent is not required.
Electronic Supplementary Material
ESM 1
(DOCX 16 kb)
Rights and permissions
About this article
Cite this article
Chen, PH., Zafar, H., Galperin-Aizenberg, M. et al. Integrating Natural Language Processing and Machine Learning Algorithms to Categorize Oncologic Response in Radiology Reports. J Digit Imaging 31, 178–184 (2018). https://doi.org/10.1007/s10278-017-0027-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-017-0027-x