Abstract
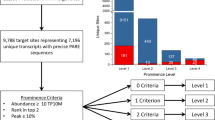
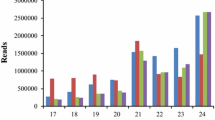
We aim to identify conserved and dehydration responsive microRNAs (miRNAs) in Hordeum vulgare (barley). A total of 28 new barley miRNAs belonging to 18 distinct miRNA families were identified. Detailed nucleotide analyses revealed that barley pre-miRNAs are in the range of 46–114 nucleotides with average of 77.14. Using 28 newly detected miRNAs as queries, 445 potential target mRNAs were predicted. The predicted miRNAs were differentially expressed and some of them behaved similarly in leaf and root tissues upon stress treatment. Hvu-MIR156, Hvu-MIR166, Hvu-MIR171, and Hvu-MIR408 were detected as dehydration stress-responsive barley miRNAs. To discover target transcripts of barley miRNAs a modified 5′ RLM-RACE was performed and seven cleaved miRNA transcripts were retrieved from drought stressed leaf samples. In silico analysis indicated 15 potential EST targets. Measurement of expression levels showed a positive correlation between levels of miRNA expression and suppression of their target mRNA transcripts in dehydration-stress-treated barley.




Similar content being viewed by others
Abbreviations
- ΔG:
-
Folding free energies
- AGO1:
-
Argonaute-1
- ARF:
-
Auxin response transcription factor
- DCL1:
-
Dicer-like 1
- GSS:
-
Genome survey sequence
- miRNA:
-
Micro-RNA
- RT:
-
Real time
- EST:
-
Expressed sequence tag
- EREBPs:
-
Ethylene-responsive element binding proteins
- MFE:
-
Minimal folding free energy
- MFEI:
-
Minimal folding free energy index
- miRNA*:
-
miRNA star strand
- NCBI:
-
National center for biotechnology information
- nt:
-
Nucleotide(s)
- pre-miRNA:
-
MicroRNA precursor
- pri-miRNAs:
-
MicroRNA primary
- RISC:
-
RNA-induced silencing complex
- 5′ RLM-RACE:
-
RNA ligase-mediated 5′ rapid amplification of cDNA ends
- qRT-PCR:
-
Quantitative real-time PCR
- SBP:
-
Squamosa promoter binding proteins
- SCL6:
-
Scarecrow-like transcription factor 6
- Ser:
-
Serine
- SPL:
-
SBP-like proteins
- Thr:
-
Threonine
- ORF:
-
Open reading frame
References
Achard P, Herr A, Baulcombe DC, Harberd NP (2004) Modulation of floral development by a gibberellin-regulated microRNA. Development 131:3357–3365
Altschul S, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Arenas-Huertero C, Pérez B, Rabanal F, Blanco-Melo D, De la Rosa C, Estrada-Navarrete G, Sanchez F, Covarrubias AA, Reyes JL (2009) Conserved and novel miRNAs in the legume Phaseolus vulgaris in response to stress. Plant Mol Biol 70:385–401
Bao N, Lye KW, Barton MK (2004) MicroRNA binding sites in Arabidopsis class IIIHD-ZIP mRNAs are required for methylation of the template chromosome. Dev Cell 7:653–662
Bartel D (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Bartel B, Bartel DP (2003) MicroRNAs: at the root of plant development? Plant Physiol 132:709–771
Bonnet E, Wuyts J, Rouze P, Van de Peer Y (2004) Detection of 91 potential in plant conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc Natl Acad Sci U S A 101:11511–11516
Carrington JC, Ambros V (2003) Role of microRNAs in plant and animal development. Science 301:336–338
Chen J, Li WX, Xie D, Peng JR, Ding SW (2004) Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of microrna in host gene expression. Plant Cell 16:1302–1313
Covarrubias AA, Reyes JL (2010) Post-transcriptional gene regulation of salinity and drought responses by plant microRNAs. Plant Cell Environ 33(4):481–489
Dryanova A, Zakharov A, Gulick PJ (2008) Data mining for miRNAs and their targets in the Triticeae. Genome 51:433–443
Ergen NZ, Dinler G, Shearman RC, Budak H (2007). Identifying, cloning and structural analysis of differentially expressed genes upon Puccinia infection of Festuca rubra var. rubra. Gene:393(1-2):145–52
Ergen NZ, Thimmapuram J, Bohnert HJ, Budak H (2009) Transcriptome pathways unique to dehydration tolerant relatives of modern wheat. Funct Integr Genomics 9(3):377–396
Guo H, Xie Q, Fei JF, Chua NH (2005) MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17:1376–1386
Guo Q, Xiang AL, Yang Q, Yang ZM (2007) Bioinformatic identification of microRNAs and their target genes from Solanum tuberosum expressed sequence tags. Chin Sci Bull 52:2380–2389
Jones-Rhoades M, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14:787–799
Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 57:19–53
Kasschau K, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, Carrington JC (2003) P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA unction. Dev Cell 4:205–217
Kim J, Jung JH, Reyes JL, Kim YS, Kim SY, Chung KS, Kim JA, Lee M, Lee Y, Kim VN, Chua NH, Park CM (2005) microRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J 42:84–94
Laufs P, Peaucelle A, Morin H, Traas J (2004) MicroRNA regulation of theCUC genes is required for boundary size control in Arabidopsis meristems. Development 131:4311–4322
Li Y, Li W, Jin YX (2005) Computational identification of novel family members of microRNA genes in Arabidopsis thaliana and Oryza sativa. Acta Biochim Biophys Sin (Shanghai) 37:75–87
Li WX, Oono Y, Zhu J, He XJ, Wu JM, Iida K, Lu XY, Cui X, Jin H, Zhu JK (2008) The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20:2238–2251
Liu H, Tian X, Li YJ, Wu CA, Zheng CC (2008) Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14:836–843
Llave C, Xie ZX, Kasschau KD, Carrington JC (2002) Cleavage of scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297:2053–2056
Lu SY, Chiang VL (2008) Stress-responsive microRNAs in Populus. Plant J 55:131–151
Lu S, Sun YH, Shi R, Clark C, Li L, Chiang VL (2005) Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 17:2186–2203
Mallory A, Dugas DV, Bartel DP, Bartel B (2004) MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol 14:1035–1046
Moxon S Jr, Szittya G, Schwach F, Rusholme Pilcher RL, Moulton V, Dalmay T (2008) Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res 18:1602–1609
Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425:257–263
Phillips JR, Dalmay T, Bartel D (2007) The role of small RNAs in abiotic stress. FEBS Lett 581(19):3592–3597
Reinhart B, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Genes Dev 16:1616–1626
Rhoades M, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110:513–520
Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8:517–527
Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P (2008) The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol Biol 67:183–195
Song C, Fang J, Li X, Liu H, Chao CT (2009) Identification and characterization of 27 conserved microRNAs in citrus. Planta 230:671
Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16:2001–2019
Trindade I, Capitão C, Dalmay T, Fevereiro MP, Santos DM (2010) miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta 231(3):705–716
Unver T, Budak H (2009) Conserved microRNAs and their targets in model grass species Brachypodium distachyon. Planta 230:659–669
Unver T, Namuth-Covert D, Budak H (2009) Review of current methodological approaches for characterizing microRNAs in plants. International Journal of Plant Genomics 2009:262463
Unver T, Bakar M, Shearman RC, Budak H (2010) Genome-wide profiling and analysis of Festuca arundinacea miRNAs and transcriptomes in response to foliar glyphosate application. Mol Gen Genomics 283:397–413
Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3:1–12
Wang J, Wang LJ, Mao YB, Cai WJ, Xue HW, Chen XY (2005) Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 17:2204–2216
Wei B, Cai T, Zhang R, Li A, Huo N, Li S, Gu QY, Vogel J, Jia J, Qi Y, Mao L (2009) Novel microRNAs uncovered by deep sequencing of small RNA transcriptomes in bread wheat (Triticum aestivum L.) and Brachypodium distachyon (L.) Beauv. Funct Integr Genomics 9:499–511
Wichadakul D, Mhuanthong W (2009) MicroPC (μPC): a comprehensive resource for predicting and comparing plant microRNAs. BMC Genomics 10:366
Wu G, Poethig RS (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133:3539–3547
Xie F, Huang SQ, Guo K, Xiang AL, Zhu YY, Nie L, Yang ZM (2007) Computational identification of novel microRNAs and targets in Brassica napus. FEBS Lett 581:1464–1474
Yin Z, Li C, Han X, Shen F (2008) Identification of conserved microRNAs and their target genes in tomato (Lycopersicon esculentum). Gene 414:60–66
Zhang B, Pan XP, Wang QL, Cobb GP, Anderson TA (2005) Identification and characterization of new plant microRNAs using EST analysis. Cell Res 15:336–360
Zhang B, Pan XP, Anderson TA (2006a) Identification of 188 conserved maize microRNAs and their targets. FEBS Lett 580:3753–3762
Zhang B, Pan XP, Cobb GP, Anderson TA (2006b) Plant microRNA: a small regulatory molecule with big impact. Dev Biol 289:3–16
Zhang B, Wang QL, Pan XP (2007a) MicroRNAs and their regulatory roles in animals and plants. J Cell Physiol 210:279–289
Zhang B, Wang QL, Wang K, Pan X, Liu F, Gou T, Cobb GP, Anderson TA (2007b) Identification of cotton microRNAs and their targets. Gene 397:26–37
Zhang B, Pan X, Stellwag EJ (2008) Identification of soybean microRNAs and their targets. Planta 229:161–182
Zhao B, Liang R, Ge L, Li W, Xiao H, Lin H, Ruan K, Jin Y (2007) Identification of drought-induced microRNAs in rice. Biochem Biophys Res Commun 354:585–590
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415
Author information
Authors and Affiliations
Corresponding author
Additional information
Melda Kantar and Turgay Unver should be regarded as joint first authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1
Pre-miRNA sequences of predicted barley miRNAs (DOC 66 kb)
Supplementary Table 2
Computationally identified target ESTs for H. vulgare miRNAs (DOC 45 kb)
Supplementary Table 3
List of primers used for quantification and validation of computer-based H. vulgare miRNAs (DOC 38 kb)
Supplementary Table 4
In silico found miRNA targets that were intended to be amplified in RLM-RACE reactions. The green part indicates in silico miRNA cleavage site, yellow parts indicate reverse primers (DOC 25 kb)
Supplementary Table 5
miRNA cleaved mRNA sequences retrieved with RLM-RACE experiments (DOC 32 kb)
Supplementary Table 6
Mean quantification values and corresponding standard deviations of triplicates for miRNA and target real-time PCRs (DOC 90 kb)
Supplementary Figure 1
Sequence alignment of RACE sequences with computationally proposed Hvu-miRNAs (DOC 27 kb)
Supplementary Figure 2
Sequence alignment of RACE sequences with known miRNAs from different plant species. * indicates reverse complimentary miRNA (DOC 26 kb)
Rights and permissions
About this article
Cite this article
Kantar, M., Unver, T. & Budak, H. Regulation of barley miRNAs upon dehydration stress correlated with target gene expression. Funct Integr Genomics 10, 493–507 (2010). https://doi.org/10.1007/s10142-010-0181-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-010-0181-4