Abstract
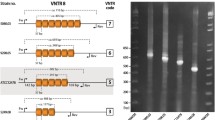
Multiple locus variable number of tandem repeats (VNTR) analysis (MLVA) has been shown to provide a high level of information for epidemiological investigations and the follow-up of Pseudomonas aeruginosa chronic infection. In the present study, an automatized MLVA assay has been developed for the analysis of 16 VNTRs in two multiplex polymerase chain reactions (PCRs), followed by capillary electrophoresis. The result in the form of a code is directly usable for clustering analyses. This MLVA-16Orsay scheme was applied to the genotyping of 83 isolates from eight cystic fibrosis patients, demonstrating that the same genotype persisted during eight years of chronic infection in the majority of cases. Comparison with pulsed-field gel electrophoresis (PFGE) analysis showed that both methods were congruent, MLVA providing, in some cases, additional informativity. The evolution of strains during long-term infection was revealed by the presence of VNTR variants.




Similar content being viewed by others
References
Saiman L, Siegel J (2004) Infection control in cystic fibrosis. Clin Microbiol Rev 17(1):57–71
Lipuma JJ (2010) The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev 23(2):299–323. doi:10.1128/CMR.00068-09
Drenkard E, Ausubel FM (2002) Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416(6882):740–743. doi:10.1038/416740a
Li Z, Kosorok MR, Farrell PM, Laxova A, West SE, Green CG, Collins J, Rock MJ, Splaingard ML (2005) Longitudinal development of mucoid Pseudomonas aeruginosa infection and lung disease progression in children with cystic fibrosis. JAMA 293(5):581–588. doi:10.1001/jama.293.5.581
Denamur E, Picard B, Decoux G, Denis JB, Elion J (1993) The absence of correlation between allozyme and rrn RFLP analysis indicates a high gene flow rate within human clinical Pseudomonas aeruginosa isolates. FEMS Microbiol Lett 110(3):275–280
Picard B, Denamur E, Barakat A, Elion J, Goullet P (1994) Genetic heterogeneity of Pseudomonas aeruginosa clinical isolates revealed by esterase electrophoretic polymorphism and restriction fragment length polymorphism of the ribosomal RNA gene region. J Med Microbiol 40(5):313–322
Römling U, Fiedler B, Bosshammer J, Grothues D, Greipel J, von der Hardt H, Tümmler B (1994) Epidemiology of chronic Pseudomonas aeruginosa infections in cystic fibrosis. J Infect Dis 170(6):1616–1621
Vu-Thien H, Corbineau G, Hormigos K, Fauroux B, Corvol H, Clément A, Vergnaud G, Pourcel C (2007) Multiple-locus variable-number tandem-repeat analysis for longitudinal survey of sources of Pseudomonas aeruginosa infection in cystic fibrosis patients. J Clin Microbiol 45(10):3175–3183. doi:10.1128/JCM.00702-07
Wiehlmann L, Wagner G, Cramer N, Siebert B, Gudowius P, Morales G, Köhler T, van Delden C, Weinel C, Slickers P, Tümmler B (2007) Population structure of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 104(19):8101–8106. doi:10.1073/pnas.0609213104
Speert DP, Campbell ME (1987) Hospital epidemiology of Pseudomonas aeruginosa from patients with cystic fibrosis. J Hosp Infect 9(1):11–21
Mahenthiralingam E, Campbell ME, Foster J, Lam JS, Speert DP (1996) Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J Clin Microbiol 34(5):1129–1135
Johnson JK, Arduino SM, Stine OC, Johnson JA, Harris AD (2007) Multilocus sequence typing compared to pulsed-field gel electrophoresis for molecular typing of Pseudomonas aeruginosa. J Clin Microbiol 45(11):3707–3712. doi:10.1128/JCM.00560-07
Curran B, Jonas D, Grundmann H, Pitt T, Dowson CG (2004) Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J Clin Microbiol 42(12):5644–5649. doi:10.1128/JCM.42.12.5644-5649.2004
García-Castillo M, Del Campo R, Morosini MI, Riera E, Cabot G, Willems R, van Mansfeld R, Oliver A, Cantón R (2011) Wide dispersion of ST175 clone despite high genetic diversity of carbapenem-nonsusceptible Pseudomonas aeruginosa clinical strains in 16 Spanish hospitals. J Clin Microbiol 49(8):2905–2910. doi:10.1128/JCM.00753-11
Vergnaud G, Pourcel C (2009) Multiple locus variable number of tandem repeats analysis. Methods Mol Biol 551:141–158
Onteniente L, Brisse S, Tassios PT, Vergnaud G (2003) Evaluation of the polymorphisms associated with tandem repeats for Pseudomonas aeruginosa strain typing. J Clin Microbiol 41(11):4991–4997
Turton JF, Turton SE, Yearwood L, Yarde S, Kaufmann ME, Pitt TL (2010) Evaluation of a nine-locus variable-number tandem-repeat scheme for typing of Pseudomonas aeruginosa. Clin Microbiol Infect 16(8):1111–1116. doi:10.1111/j.1469-0691.2009.03049.x
Van der Bij AK, Van Mansfeld R, Peirano G, Goessens WH, Severin JA, Pitout JD, Willems R, Van Westreenen M (2011) First outbreak of VIM-2 metallo-beta-lactamase-producing Pseudomonas aeruginosa in The Netherlands: microbiology, epidemiology and clinical outcomes. Int J Antimicrob Agents 37(6):513–518. doi:10.1016/j.ijantimicag.2011.02.010
van Mansfeld R, Jongerden I, Bootsma M, Buiting A, Bonten M, Willems R (2010) The population genetics of Pseudomonas aeruginosa isolates from different patient populations exhibits high-level host specificity. PLoS One 5(10):e13482. doi:10.1371/journal.pone.0013482
Bingen E, Bonacorsi S, Rohrlich P, Duval M, Lhopital S, Brahimi N, Vilmer E, Goering RV (1996) Molecular epidemiology provides evidence of genotypic heterogeneity of multidrug-resistant Pseudomonas aeruginosa serotype O:12 outbreak isolates from a pediatric hospital. J Clin Microbiol 34(12):3226–3229
Vu-Thien H, Moissenet D, Valcin M, Dulot C, Tournier G, Garbarg-Chenon A (1996) Molecular epidemiology of Burkholderia cepacia, Stenotrophomonas maltophilia, and Alcaligenes xylosoxidans in a cystic fibrosis center. Eur J Clin Microbiol Infect Dis 15(11):876–879
Grundmann H, Schneider C, Hartung D, Daschner FD, Pitt TL (1995) Discriminatory power of three DNA-based typing techniques for Pseudomonas aeruginosa. J Clin Microbiol 33(3):528–534
Sobral D, Le Cann P, Gerard A, Jarraud S, Lebeau B, Loisy-Hamon F, Vergnaud G, Pourcel C (2011) High-throughput typing method to identify a non-outbreak-involved Legionella pneumophila strain colonizing the entire water supply system in the town of Rennes, France. Appl Environ Microbiol 77(19):6899–6907. doi:10.1128/AEM.05556-11
Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV (2000) Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406(6799):959–964. doi:10.1038/35023079
Yang L, Haagensen JA, Jelsbak L, Johansen HK, Sternberg C, Høiby N, Molin S (2008) In situ growth rates and biofilm development of Pseudomonas aeruginosa populations in chronic lung infections. J Bacteriol 190(8):2767–2776. doi:10.1128/JB.01581-07
Ballard LW, Adams PS, Bao Y, Bartley D, Bintzler D, Kasch L, Petukhova L, Rosato C (2002) Strategies for genotyping: effectiveness of tailing primers to increase accuracy in short tandem repeat determinations. J Biomol Tech 13(1):20–29
Waters RC, O’Toole PW, Ryan KA (2007) The FliK protein and flagellar hook-length control. Protein Sci 16(5):769–780. doi:10.1110/ps.072785407
Vogler AJ, Keys CE, Allender C, Bailey I, Girard J, Pearson T, Smith KL, Wagner DM, Keim P (2007) Mutations, mutation rates, and evolution at the hypervariable VNTR loci of Yersinia pestis. Mutat Res 616(1–2):145–158. doi:10.1016/j.mrfmmm.2006.11.007
Noller AC, McEllistrem MC, Shutt KA, Harrison LH (2006) Locus-specific mutational events in a multilocus variable-number tandem repeat analysis of Escherichia coli O157:H7. J Clin Microbiol 44(2):374–377. doi:10.1128/JCM.44.2.374-377.2006
Price EP, Hornstra HM, Limmathurotsakul D, Max TL, Sarovich DS, Vogler AJ, Dale JL, Ginther JL, Leadem B, Colman RE, Foster JT, Tuanyok A, Wagner DM, Peacock SJ, Pearson T, Keim P (2010) Within-host evolution of Burkholderia pseudomallei in four cases of acute melioidosis. PLoS Pathog 6(1):e1000725. doi:10.1371/journal.ppat.1000725
Acknowledgments
D.S., F.L.-H., and B.L. are employees of Ceeram and hold stocks. This study was performed with the support of the association Vaincre La Mucoviscidose (grant no. RC0630). The development of tools for the surveillance of bacterial pathogens is supported by the French Direction Générale de l’Armement.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sobral, D., Mariani-Kurkdjian, P., Bingen, E. et al. A new highly discriminatory multiplex capillary-based MLVA assay as a tool for the epidemiological survey of Pseudomonas aeruginosa in cystic fibrosis patients. Eur J Clin Microbiol Infect Dis 31, 2247–2256 (2012). https://doi.org/10.1007/s10096-012-1562-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-012-1562-5