Abstract
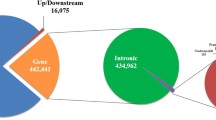
Since the completion of the bovine sequencing projects, a substantial number of genetic variations such as single nucleotide polymorphisms have become available across the cattle genome. Recently, cataloguing such genetic variations has been accelerated using massively parallel sequencing technology. However, most of the recent studies have been concentrated on European Bos taurus cattle breeds, resulting in a severe lack of knowledge for valuable native cattle genetic resources worldwide. Here, we present the first whole-genome sequencing results for an endangered Korean native cattle breed, Chikso, using the Illumina HiSeq 2,000 sequencing platform. The genome of a Chikso bull was sequenced to approximately 25.3-fold coverage with 98.8% of the bovine reference genome sequence (UMD 3.1) covered. In total, 5,874,026 single nucleotide polymorphisms and 551,363 insertion/deletions were identified across all 29 autosomes and the X-chromosome, of which 45% and 75% were previously unknown, respectively. Most of the variations (92.7% of single nucleotide polymorphisms and 92.9% of insertion/deletions) were located in intergenic and intron regions. A total of 16,273 single nucleotide polymorphisms causing missense mutations were detected in 7,111 genes throughout the genome, which could potentially contribute to variation in economically important traits in Chikso. This study provides a valuable resource for further investigations of the genetic mechanisms underlying traits of interest in cattle, and for the development of improved genomics-based breeding tools.
Similar content being viewed by others
References
Abecasis, G.R., Auton, A., Brooks, L.D., DePristo, M.A., Durbin, R.M., Handsaker, R.E., Kang, H.M., Marth, G.T., and McVean, G.A. (2012). An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65.
Bovine Genome Sequencing Analysis Consortium, Elsik, C.G., Tellam, R.L., Worley, K.C., Gibbs, R.A., Muzny, D.M., Weinstock, G.M., Adelson, D.L., Eichler, E.E., Elnitski, L., et al. (2009). The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science 324, 522–528.
Bovine HapMap Consortium, Gibbs, R.A., Taylor, J.F., Van Tassell, C.P., Barendse, W., Eversole, K.A., Gill, C.A., Green, R.D., Hamernik, D.L., Kappes, S.M., et al. (2009). Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science 324, 528–532.
Choi, T.J. (2009). Establishment of phylogenomic characteristics for Korean traditional cattle breeds (Hanwoo, Korean brindle and black). Doctoral Thesis. Jeon-buk National University, Republic of Korea.
Dadi, H., Lee, S.H., Jung, K.S., Choi, J.W., Ko, M.S., Han, Y.J., Kim, J.J., and Kim, K.S. (2012). Effect of population reduction on mtDNA diversity and demographic history of Korean Cattle populations. AJAS 25, 1223–1228.
de Souza, F.R., Chiquitelli, M.G., da Fonseca, L.F., Cardoso, D.F., da Silva Fonseca, P.D., de Camargo, G.M., Gil, F.M., Boligon, A. A., Tonhati, H., Mercadante, M.E., et al. (2012). Associations of FASN gene polymorphisms with economical traits in Nellore cattle (Bos primigenius indicus). Mol. Biol. Rep. 39, 10097–10104.
Eck, S.H., Benet-Pages, A., Flisikowski, K., Meitinger, T., Fries, R., and Strom, T.M. (2009). Whole genome sequencing of a single Bos taurus animal for single nucleotide polymorphism discovery. Genome Biol. 10, R82.
FAO (Food and Agriculture Organization). (2012). Domestic Animal Diversity Information Service (DAD-IS). http://dad.fao.org/ Accessed December 20, 2012.
Flicek, P., Amode, M.R., Barrell, D., Beal, K., Brent, S., Carvalho-Silva, D., Clapham, P., Coates, G., Fairley, S., Fitzgerald, S., et al. (2012). Ensembl 2012. Nucleic Acids Res. 40, D84–90.
Grant, J.R., Arantes, A.S., Liao, X., and Stothard, P. (2011). Indepth annotation of SNPs arising from resequencing projects using NGS-SNP. Bioinformatics 27, 2300–2301.
Jo, C., Cho, S.H., Chang, J., and Nam, K.C. (2012). Keys to production and processing of Hanwoo beef: a perspective of tradition and science. Animal Frontiers 2, 32–38.
Kawahara-Miki, R., Tsuda, K., Shiwa, Y., Arai-Kichise, Y., Matsumoto, T., Kanesaki, Y., Oda, S., Ebihara, S., Yajima, S., Yoshikawa, H., et al. (2011). Whole-genome resequencing shows numerous genes with nonsynonymous SNPs in the Japanese native cattle Kuchinoshima-Ushi. BMC Genomics 12, 103.
Kim, K.H., Lee, J.H., Lee, S.C., Park, W.Y., Oh, Y.G., Kang, S.W., and Ko, Y.D. (2005). The optimal TDN levels of concentrates and slaughter age in Hanwoo steers. J. Anim. Sci. Technol. 47, 731–744.
Klungland, H., Vage, D.I., Gomez-Raya, L., Adalsteinsson, S., and Lien, S. (1995). The role of melanocyte-stimulating hormone (MSH) receptor in bovine coat color determination. Mamm. Genome 6, 636–639.
Levy, S., Sutton, G., Ng, P.C., Feuk, L., Halpern, A.L., Walenz, B.P., Axelrod, N., Huang, J., Kirkness, E.F., Denisov, G., et al. (2007). The diploid genome sequence of an individual human. PLoS Biol. 5, e254.
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760.
Li, W.H., Yi, S., and Makova, K. (2002). Male-driven evolution. Curr. Opin. Genet. Dev. 12, 650–656.
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., Marth, G., Abecasis, G., Durbin, R., and Genome Project Data Processing, S. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079.
Magrane, M., and Consortium, U. (2011). UniProt Knowledgebase: a hub of integrated protein data. Database 2011, bar009.
Makova, K.D., and Li, W.H. (2002). Strong male-driven evolution of DNA sequences in humans and apes. Nature 416, 624–626.
NIAS (National Institute of Animal Science). (2012). The status of local livestock breeds in Korea, registered in DAD-IS. http://www.nias.go.kr/ Accessed December 20, 2012.
Sayers, E.W., Barrett, T., Benson, D.A., Bolton, E., Bryant, S.H., Canese, K., Chetvernin, V., Church, D.M., Dicuccio, M., Federhen, S., et al. (2012). Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 40, D13–25.
Seo, K., Mohanty, T.R., Choi, T., and Hwang, I. (2007). Biology of epidermal and hair pigmentation in cattle: a mini-review. Vet. Dermatol. 18, 392–400.
Smith, S.B., Gill, C.A., Lunt, D.K., and Brooks, M.A. (2009). Regulation of fat and fatty acid composition in beef cattle. AJAS 22, 1225–1233.
Stothard, P., Choi, J.W., Basu, U., Sumner-Thomson, J.M., Meng, Y., Liao, X., and Moore, S.S. (2011). Whole genome resequencing of black Angus and Holstein cattle for SNP and CNV discovery. BMC Genomics 12, 559.
Yeon, S.H., Lee, S.H., Choi, B.H., Lee, H.J., Jang, G.W., Lee, K.T., Kim, K.H., Lee, J.H., and Chung, H.Y. (2013). Genetic variation of FASN is associated with fatty acid composition of Hanwoo. Meat Sci. 94, 133–138.
Zhang, S., Knight, T.J., Reecy, J.M., Wheeler, T.L., Shackelford, S.D., Cundiff, L.V., and Beitz, D.C. (2010). Associations of polymorphisms in the promoter I of bovine acetyl-CoA carboxylase-alpha gene with beef fatty acid composition. Anim. Genet. 41, 417–420.
Zimin, A.V., Delcher, A.L., Florea, L., Kelley, D.R., Schatz, M.C., Puiu, D., Hanrahan, F., Pertea, G., Van Tassell, C.P., Sonstegard, T.S., et al. (2009). A whole-genome assembly of the domestic cow, Bos taurus. Genome Biol. 10, R42.
Author information
Authors and Affiliations
Corresponding authors
About this article
Cite this article
Choi, JW., Liao, X., Park, S. et al. Massively parallel sequencing of Chikso (Korean brindle cattle) to discover genome-wide SNPs and InDels. Mol Cells 36, 203–211 (2013). https://doi.org/10.1007/s10059-013-2347-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10059-013-2347-0