Abstract
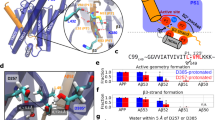
The formation of β-amyloid peptide (Aβ) is initiated from cleavage of amyloid precursor protein (APP) by a family of protease, α-, β-, and γ-secretase. Sub W, a substrate peptide, consists of 10 amino acids, which are adjacent to the β-cleavage site of wild-type APP, and Sub M is Swedish mutant with double mutations on the left side of the β-cleavage site of APP. Sub W is a normal product of the metabolism of APP in the secretary pathway. Sub M is known to increase the efficiency of β-secretase activity, resulting in a more specific binding model compared to Sub W. Three-dimensional structures of Sub W and Sub M were studied by CD and NMR spectroscopy in water solution. On the basis of these structures, interaction models of β-secretase and substrate peptides were determined by molecular dynamics simulation. Four hydrogen bonds and one water-mediated interaction were formed in the docking models. In particular, the hydrogen bonding network of Sub M-BACE formed spread over the broad region of the active site of β-secretase (P5-P3′), and the side chain of P2-Asn formed a hydrogen bond specifically with the side chain of Arg235. These are more favorable to the cleavage of Sub M by β-secretase than Sub W. The two substrate peptides showed different tendency to bind to β-secretase and this information may useful for drug development to treat and prevent Alzheimer’s disease.
Similar content being viewed by others
References
Bax, A., and Davis, D.G. (1985a). MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 65, 355–360.
Bax, A., and Davis, D.G. (1985b). Practical aspects of two-dimensional transverse NOE spectroscopy. J. Magn. Reson. 63, 207–213.
Brik, A., and Wong, C.H. (2003). HIV-1 protease: mechanism and drug discovery. Org. Biomol. Chem. 1, 5–14.
Buxbaum, J.D., Gandy, S.E., Cicchetti, P., Ehrlich, M.E., Czernik, A.J., Fracasso, R.P., Ramabhadran, T.V., Unterbeck, A.J., and Greengard, P. (1990). Processing of Alzheimer beta/A4 amyloid precursor protein: modulation by agents that regulate protein phosphorylation. Proc. Natl. Acad. Sei. USA 87, 6003–6006.
Citron, M. (2004). β-Secretase inhibition for the treatment of Alzheimer’s disease promise and challenge. Trends Pharmacol. Sci. 25, 92–97.
Clore, G.M., Gronenborn, A.M., Nilges, M., and Ryan, C.A. (1987). Three-dimensional structure of potato carboxypeptidase inhibitor in solution. A study using nuclear magnetic resonance, distance geometry, and restrained molecular dynamics. Biochemistry 26, 8012–8023.
Clore, G.M., and Gronenborn, A.M. (1989). Determination of three-dimensional structures of proteins and nucleic acids in solution by nuclear magnetic resonance spectroscopy. Crit. Rev. Biochem. Mol. Biol. 24, 479–564.
Clore, G.M., and Gronenborn, AM. (1994). Structures of larger proteins, protein-ligand and protein-DNA complexes by multidimensional heteronuclear NMR. Prog. Biophys. Mol. Biol. 62, 153–184.
Cras, P., Kawai, M., Lowery, D., Gonzalez-DeWhitt, P., Greenberg, B., and Perry, G. (1991). Senile plaque neuritis in Alzheimer disease accumulate amyloid precursor protein. Proc. Natl. Acad. Sci. USA 88, 7552–7556.
Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. (1995). NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293.
Derome, A.E., and Williamson, M.P. (1990). Rapid-pulsing artifacts in double-quantum-filtered COSY. J. Magn. Reson. 88, 177–185.
Esler, W.P., and Wolfe, M.S. (2001). A portrait of Alzheimer secretases-New features and familiar faces. Science 293, 1449–1454.
Goddard, T.D., and Kneller, D.G. (2001). SPARKY3, University of California, San Francisco.
Goodsell, D.S., Morris, G.M., and Olson, A.J. (1996). Automated docking of flexible ligands: applications of AutoDock. J. Mol. Recognit. 9, 1–5.
Gregor, W.G. (2006). Dissertation: identification of a BACE dimer and characterization of its biochemical and enzymatic properties. Ludwig-Maximilians-Universität München, Medizinische Fakultät.
Grüninger-Leitch, F., Schlatter, D., Küng, E., Nelböck, P., and Döbeli, H. (2002). Substrate and inhibitor profile of BACE (β-secretase) and comparison with other mammalian aspartic proteases. J. Biol. Chem. 277, 4687–4693.
Haass, C. (2004). Take five-BACE and the γ-secretase quartet conduct Alzheimer’s amyloid β-peptide generation. EMBO J. 23, 484–488.
Haass, C., Lemere, C.A., Capell, A., Citron, M., Seubert, P., Schenk, D., Lannfelt, L, and Selkoe, D.J. (1995). The Swedish mutation causes early-onset Alzheimer’s disease by β-secretase cleavage within the secretory pathway. Nat. Med. 1, 1291–1296.
Han, K.D., Park, S.J., Jang, S.B., and Lee, B.J. (2008). Backbone 1H, 15N, and 13C resonance assignments and secondary-structure of the conserved hypothetical protein HP0892 of Helicobacter pylori. Mol. Cells 25, 138–141.
Hong, L, Koelsch, G., Lin, X., Wu, S., Terzyan, S., Ghosh, A.K., Zhang, X.C, and Tang, J. (2000). Structure of the protease domain of Memapsin 2 (β-secretase) complexed with inhibitor. Science 290, 150–153.
Hu, B., Xiong, B., Qiu, B.Y., Li, X., Yu, H.P., Xiao, K., Wang, X., Li, J., and Shen, J.K. (2006). Construction of a small peptide library related to inhibitor OM99-2 and its structure-activity relationship to β-secretase. Acta Pharmacol. Sin. 27, 1586–1593.
Kagan, B.L., Azimov, R., and Azimova, R. (2004). Amyloid peptide channels. J. Membrane Biol. 202, 1–10.
Kitazume, S., Tachida, Y., Oka, R., Kotani, N., Ogawa, K., Suzuki, M., Dohmae, N., Takio, K., Saido, T.C., and Hashimoto, Y. (2003). Characterization of α2,6-sialyltransferase cleavage by Alzheimer’s β-secretase (BACE1). J. Biol. Chem. 278, 14865–14871.
Kuszewski, J., Nilges, M., and Brünger, A.T. (1992). Sampling and efficiency of metric matrix distance geometry: a novel partial metization algorithm. J. Biomol. NMR 2, 33–56.
Laskowski, R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. (1993). PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283.
Li, R., Lindholm, K., Yang, L.B., Yue, X., Citron, M., Yan, R., Beach, T., Sue, L., Sabbagh, M., Cai, H., et al. (2004). Amyloid β peptide load is correlated with increased β-secretase activity in sporadic Alzheimer’s disease patients. Proc. Natl. Acad. Sei. USA 101, 3632–3637.
Marion, D., and Wüthrich, K. (1983). Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H-1H spin-spin coupling constants in proteins. Biochem. Biophys. Res. Commun. 113, 967–974.
Martin, B.L, Schrader-Fischer, G., Busciglio, J., Duke, M., Paganetti, P., and Yankner, B.A. (1995). Intracellular accumulation of β-amyloid in cells expressing the Swedish mutant amyloid precursor protein. J. Biol. Mol. 270, 26727–26730.
Na, C.H., Jeon, S.H., Zhang, G., Olson, G.L, and Chae, C.B. (2007). Inhibition of Amyloid β-peptide production by blockage of β-secretase cleavage site of amyloid precursor protein. J. Neurochem. 101, 1583–1595.
Nilges, M., Clore, G.M., and Gronenbom, A.M. (1988). Determination of three-dimensional structures of proteins from interproton distance data by hybrid distance geometry-dynamical simulated annealing calculations. FEBS Lett. 229, 317–324.
Numan, J., and Small, D.H. (2000). Regulation of APP cleavage by α-, β-, and γ-secretases. FEBS Lett. 483, 6–10.
Patel, S., Vuillard, L, Cleasby, A, Murray, C.W., and Yon, J. (2004). Apo and inhibitor complex structures of BACE (β-secretase). J. Mol. Biol. 343, 407–16.
Rajamani, R., and Reynolds, C.H. (2004). Modeling the protonation states of the catalytic aspartates in β-secretase. J. Med. Chem. 47, 5159–5166.
Roggo, S. (2002). Inhibition of BACE, a promising approach to Alzheimer’s disease therapy. Curr. Top. Med. Chem. 2, 359–370.
Sauder, J.M., Arthur, J.W., and Dunbrack, R.L. Jr. (2000). Modeling of substrate specificity of the Alzheimer’s disease amyloid precursor protein β-secretase. J. Mol. Biol. 300, 241–248.
Schmechel, A., Strauss, M., Schlicksupp, A., Pipkorn, R., Haass, C., Bayer, T.A., and Multhaup G. (2004). Human BACE forms dimers and colocalizes with APP. J. Biol. Chem. 279, 39710–39717.
Suguna, K., Padlan, E.A, Smith, C.W., Carlson, W.D., and Davies, D.R. (1987). Binding of a reduced peptide inhibitor to the aspartic proteinase from Rhizopus chinensis: implications for a mechanism of action. Proc. Natl. Acad. Sci. USA 84, 7009–7013.
Tang, B.L. (2005). Alzheimer’s disease: channeling APP to non-amyloidogenic processing. Biochem. Biophys. Res. Commun. 331, 375–378.
Toulokhonova, L., Metzler, W.J., Witmer, M.R., Copeland, R.A, and Marcinkeviciene, J. (2003). Kinetic studies on β-Site amyloid precursor protein-cleaving enzyme (BACE). J. Biol. Chem. 278, 4582–4589.
Turner, R.T., 3rd., Hong, L, Koelsch, G., Ghosh, A.K., and Tang, J. (2005). Structural locations and functional roles of new subsites S5, S6, and S7 in me maps in 2 (β-secretase). Biochemistry 44, 105–112.
Vassar, R., Bennett, B.D., Babu-Khan, S., Kahn, S., Mendiaz, E.A., Denis, P., Teplow, D.B., Ross, S., Amarante, P., Loeloff, R., et al. (1999). β-Secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science 286, 735–741.
Wüthrich, K. (1986). NMR of proteins and nucleic acids, Wiley, New York.
Wüthrich, K, Billeter, M., and Braun, W. (1983). Pseudo-structures for the 20 common amino acids for use in studies of protein conformations by measurements of intramolecular proton-proton distance constraints with nuclear magnetic resonance. J. Mol. Biol. 169, 949–961.
Xu, Y, Shen, J., Luo, X., Zhu, W., Chen, K, Ma, J., and Jiang, H. (2005). Conformational transition of amyloid β-peptide. Proc. Natl. Acad. Sci. USA 102, 5403–5407.
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Lee, JY., Lee, SA., Kim, JK. et al. Interaction models of substrate peptides and β-secretase studied by NMR spectroscopy and molecular dynamics simulation. Mol Cells 27, 651–656 (2009). https://doi.org/10.1007/s10059-009-0086-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10059-009-0086-z