Abstract
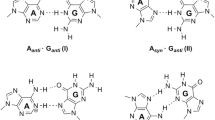
In this paper, we aim to determine whether the N7-methylation can influence the base pairing properties of guanine by promoting the formation of guanine enol-tautomers. The keto- to -enol-tautomerization of N7-methylguanine (N7mG) and its base pairing patterns with all the canonical DNA bases have been investigated at the M06-2X/6-311+G(d,p) level of density functional theory. The barrier free energy calculations reveal that N7-methylation does not promote the keto- to enol- tautomerization of guanine. The Watson-Crick-like enol-N7mG:T1 or enol-N7mG:T2 base pair similar to what is observed experimentally is found to be energetically more stable than the keto-N7mG:T base pairs. However, the keto-N7mG:C1 which is structurally similar to the canonical G:C base pair is the most stable base pair among all the base pairs studied here. Thus, our calculations predict that N7mG would pair preferably with cytosine during DNA replication but there is also a probability that it can cause mutation through mispairing with thymine, in agreement with experimental observations.




Similar content being viewed by others
Data availability
All data generated or analysed during this study are included in this published article.
References
Das PM, Singal R (2004) DNA methylation and cancer. J Clin Oncol 22(22):4632–4642
Giglia-Mari G, Zotter A, Vermeulen W (2011) DNA damage response. Cold Spring Harb Perspect Biol 3(1):a000745
Brown R, Plumb JA (2004) Demethylation of DNA by decitabine in cancer chemotherapy. Expert Rev Anticancer Ther 4(4):501–510
David SS, O'Shea VL, Kundu S (2007) Base-excision repair of oxidative DNA damage. Nature 447(7147):941–950
Egger G, Liang G, Aparicio A, Jones PA (2004) Epigenetics in human disease and prospects for epigenetic therapy. Nature 429(6990):457–463. https://doi.org/10.1038/nature02625
Feinberg AP, Ohlsson R, Henikoff S (2006) The epigenetic progenitor origin of human cancer. Nat Rev Genet 7(1):21–33. https://doi.org/10.1038/nrg1748
Helleday T, Lo J, van Gent DC, Engelward BP (2007) DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair (Amst) 6(7):923–935
Hoeijmakers JH (2009) DNA damage, aging, and cancer. N Engl J Med 361(15):1475–1485
Hussain SP, Hofseth LJ, Harris CC (2003) Radical causes of cancer. Nat Rev Cancer 3(4):276–285
Jena N (2012) DNA damage by reactive species: mechanisms, mutation and repair. J Biosci 37(3):503–517
Klaunig JE, Kamendulis LM (2004) The role of oxidative stress in carcinogenesis. Annu Rev Pharmacol Toxicol 44:239–267
Olinski R, Gackowski D, Foksinski M, Rozalski R, Roszkowski K, Jaruga P (2002) Oxidative DNA damage: assessment of the role in carcinogenesis, atherosclerosis, and acquired immunodeficiency syndrome. Free Radic Biol Med 33(2):192–200
Weidman JR, Dolinoy DC, Murphy SK, Jirtle RL (2007) Cancer susceptibility: epigenetic manifestation of environmental exposures. Cancer J 13(1):9–16
Kulis M, Esteller M (2010) DNA methylation and cancer. Advances in genetics. Elsevier, pp 27–56
Shukla P, Jena N, Mishra P (2011) Quantum theoretical study of molecular mechanisms of mutation and cancer-a review. Proc Natl Acad Sci India Sect A Phys Sci 81(part 2):79–98
Gates KS (2009) An overview of chemical processes that damage cellular DNA: spontaneous hydrolysis, alkylation, and reactions with radicals. Chem Res Toxicol 22(11):1747–1760
Halliwell B (1999) Oxygen and nitrogen are pro-carcinogens. Damage to DNA by reactive oxygen, chlorine and nitrogen species: measurement, mechanism and the effects of nutrition. Mutat Res 443(1–2):37–52
Lawley P, Brookes P (1963) Further studies on the alkylation of nucleic acids and their constituent nucleotides. Biochem J 89(1):127
Lawley P (1966) Effects of some chemical mutagens and carcinogens on nucleic acids. Prog Nucleic Acid Res Mol Biol 5:89–131
Lawley P (1989) Mutagens as carcinogens: development of current concepts. Mutat Res 213(1):3–25
Mishina Y, Duguid EM, He C (2006) Direct reversal of DNA alkylation damage. Chem Rev 106(2):215–232
Neeley WL, Essigmann JM (2006) Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem Res Toxicol 19(4):491–505
Niles JC, Wishnok JS, Tannenbaum SR (2006) Peroxynitrite-induced oxidation and nitration products of guanine and 8-oxoguanine: structures and mechanisms of product formation. Nitric Oxide 14(2):109–121
Wyatt MD, Pittman DL (2006) Methylating agents and DNA repair responses: methylated bases and sources of strand breaks. Chem Res Toxicol 19(12):1580–1594
Sonntag C (2006) Free-radical-induced DNA damage and its repair: a chemical perspective. Springer, Berlin, Heidelberg
Chen D, Meng L, Pei F, Zheng Y, Leng J (2017) A review of DNA methylation in depression. J Clin Neurosci 43:39–46
Fu D, Calvo JA, Samson LD (2012) Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat Rev Cancer 12(2):104
Meikrantz W, Bergom MA, Memisoglu A, Samson L (1998) O6-alkylguanine DNA lesions trigger apoptosis. Carcinogenesis 19(2):369–372
Robison SH, Munzer JS, Mrcp RT, Bradley WG (1987) Alzheimer's disease cells exhibit defective repair of alkylating agent—induced DNA damage. Ann Neurol 21(3):250–258
Friedberg EC, Walker GC, Siede W, Wood RD (2005) DNA repair and mutagenesis. American Society for Microbiology Press, Washington, DC
Gates KS, Nooner T, Dutta S (2004) Biologically relevant chemical reactions of N7-alkylguanine residues in DNA. Chem Res Toxicol 17(7):839–856
Singer B (1975) The chemical effects ofNucleic acid alkylation and their relation to mutagenesis and carcinogen esis. Prog Nucleic Acid Res Mol Biol 15:219–284
Singer B, Grunberger D (2012) Molecular biology of mutagens and carcinogens. Plenum Press, New York
de Vries M, van der Plaat DA, Nedeljkovic I, Verkaik-Schakel RN, Kooistra W, Amin N, van Duijn CM, Brandsma C-A, van Diemen CC, Vonk JM (2018) From blood to lung tissue: effect of cigarette smoke on DNA methylation and lung function. Respir Res 19(1):1–9
Reynolds LM, Wan M, Ding J, Taylor JR, Lohman K, Su D, Bennett BD, Porter DK, Gimple R, Pittman GS (2015) DNA methylation of the aryl hydrocarbon receptor repressor associations with cigarette smoking and subclinical atherosclerosis. Circ Cardiovasc Genet 8(5):707–716
Drabløs F, Feyzi E, Aas PA, Vaagbø CB, Kavli B, Bratlie MS, Peña-Diaz J, Otterlei M, Slupphaug G, Krokan HE (2004) Alkylation damage in DNA and RNA—repair mechanisms and medical significance. DNA Repair 3(11):1389–1407
Cai J, Zhao Y, Liu P, Xia B, Zhu Q, Wang X, Song Q, Kan H, Zhang Y (2017) Exposure to particulate air pollution during early pregnancy is associated with placental DNA methylation. Sci Total Environ 607:1103–1108
Joubert BR, Håberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, Huang Z, Hoyo C, Midttun Ø, Cupul-Uicab LA (2012) 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect 120(10):1425–1431
Zeilinger S, Kühnel B, Klopp N, Baurecht H, Kleinschmidt A, Gieger C, Weidinger S, Lattka E, Adamski J, Peters A (2013) Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS One 8(5):e63812
Waddell JA, Solimando Jr DA (2006) Carmustine, cisplatin, dacarbazine, and tamoxifen (Dartmouth regimen) for metastatic melanoma. Hosp Pharm 41(2):124–132
Cocconi G, Bella M, Calabresi F, Tonato M, Canaletti R, Boni C, Buzzi F, Ceci G, Corgna E, Costa P (1992) Treatment of metastatic malignant melanoma with dacarbazine plus tamoxifen. N Engl J Med 327(8):516–523
O'day SJ, Kim CJ, Reintgen DS (2002) Metastatic melanoma: chemotherapy to biochemotherapy. Cancer Control 9(1):31–38
Eggermont AM, Kirkwood JM (2004) Re-evaluating the role of dacarbazine in metastatic melanoma: what have we learned in 30 years? Eur J Cancer 40(12):1825–1836
Douglas JG, Margolin K (2002) The treatment of brain metastases from malignant melanoma. Seminars in oncology. Elsevier, pp 518–524
Zhang J, Stevens MFG, Bradshaw TD (2012) Temozolomide: mechanisms of action, repair and resistance. Curr Mol Pharmacol 5(1):102–114
Boysen G, Pachkowski BF, Nakamura J, Swenberg JA (2009) The formation and biological significance of N7-guanine adducts. Mutat Res 678(2):76–94
Ekanayake KS, LeBreton PR (2006) Activation barriers for DNA alkylation by carcinogenic methane diazonium ions. J Comput Chem 27(3):277–286
Sedgwick B (2004) Repairing DNA-methylation damage. Nat Rev Mol Cell Biol 5(2):148–157
Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T (2007) Repair of alkylated DNA: recent advances. DNA Repair 6(4):429–442
Boiteux S, Guillet M (2004) Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair 3(1):1–12
Dutta S, Chowdhury G, Gates KS (2007) Interstrand cross-links generated by abasic sites in duplex DNA. J Am Chem Soc 129(7):1852–1853
Greenberg MM (2014) Abasic and oxidized abasic site reactivity in DNA: enzyme inhibition, cross-linking, and nucleosome catalyzed reactions. Acc Chem Res 47(2):646–655
Lhomme J, Constant JF, Demeunynck M (1999) Abasic DNA structure, reactivity, and recognition. Biopolymers 52(2):65–83
Loeb LA, Preston BD (1986) Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet 20(1):201–230
Wilson III DM, Barsky D (2001) The major human abasic endonuclease: formation, consequences and repair of abasic lesions in DNA. Mutat Res 485(4):283–307
Citti L, Gervasi P, Turchi G, Ga B, Bianchini R (1984) The reaction of 3, 4-epoxy-1-butene with deoxyguanosine and DNA in vitro: synthesis and characterization of the main adducts. Carcinogenesis 5(1):47–52
King H, Osborne M, Brookes P (1979) The in vitro and in vivo reaction at the N7-position of guanine of the ultimate carcinogen derived from benzo [a] pyrene. Chem Biol Interact 24(3):345–353
Pujari SS, Tretyakova N (2017) Chemical biology of N5-substituted formamidopyrimidine DNA adducts. Chem Res Toxicol 30(1):434–452
Price NE, Johnson KM, Wang J, Fekry MI, Wang Y, Gates KS (2014) Interstrand DNA–DNA cross-link formation between adenine residues and abasic sites in duplex DNA. J Am Chem Soc 136(9):3483–3490
Tudek B (2003) Imidazole ring-opened DNA purines and their biological significance. J Biochem Mol Biol 36(1):12–19
Wiederholt CJ, Greenberg MM (2002) Fapy⊙ dG instructs Klenow exo-to misincorporate deoxyadenosine. J Am Chem Soc 124(25):7278–7279
Yang K, Park D, Tretyakova NY, Greenberg MM (2018) Histone tails decrease N7-methyl-2′-deoxyguanosine depurination and yield DNA–protein cross-links in nucleosome core particles and cells. Proc Natl Acad Sci 115(48):E11212–E11220
Kou Y, Koag M-C, Lee S (2015) N7 methylation alters hydrogen-bonding patterns of guanine in duplex DNA. J Am Chem Soc 137(44):14067–14070
Barone G, Fonseca Guerra C, Bickelhaupt FM (2013) B-DNA structure and stability as function of nucleic acid composition: dispersion-corrected DFT study of dinucleoside monophosphate single and double strands. ChemistryOpen 2(5–6):186–193
Šponer J, Jurecka P, Hobza P (2004) Accurate interaction energies of hydrogen-bonded nucleic acid base pairs. J Am Chem Soc 126(32):10142–10151
Fonseca Guerra C, Bickelhaupt FM (1999) Charge transfer and environment effects responsible for characteristics of DNA base pairing. Angew Chem Int Ed 38(19):2942–2945
Shukla PK, Mishra P (2013) Base pairing patterns of DNA base lesion spiroiminodihydantoin: a DFT study. Int J Quantum Chem 113(24):2600–2604
Felske LR, Lenz SA, Wetmore SD (2018) Quantum chemical studies of the structure and stability of N-methylated DNA nucleobase dimers: insights into the mutagenic base pairing of damaged DNA. J Phys Chem A 122(1):410–419
Flood A, Hubbard C, Forde G, Hill G, Gorb L, Leszczynski J (2003) Theoretical ab initio study of the effects of methylation on the nature of hydrogen bonding in A:T base pair. J Biomol Struct Dyn 21(2):297–302
Forde G, Flood A, Salter L, Hill G, Gorb L, Leszczynski J (2003) Theoretical ab initio study of the effects of methylation on structure and stability of G: C Watson-Crick base pair. J Biomol Struct Dyn 20(6):811–817
Zhao Y, Truhlar DG (2008) The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor Chem Accounts 120(1–3):215–241
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA (2009) Gaussian 09 C. 01. Gaussian Inc, Wallingford CT
Mennucci B, Tomasi J (1997) Continuum solvation models: a new approach to the problem of solute’s charge distribution and cavity boundaries. J Chem Phys 106(12):5151–5158
Dennington R, Keith T, Millam J (2009) GaussView, version 5
Forde GK, Forde AE, Hill G, Ford A, Nazario A, Leszczynski J (2006) Comprehensive study of the effects of methylation on tautomeric equilibria of nucleic acid bases. J Phys Chem B 110(31):15564–15571
Yanson I, Teplitsky A, Sukhodub L (1979) Experimental studies of molecular interactions between nitrogen bases of nucleic acids. Biopolymers 18(5):1149–1170
Rejnek J, Hobza P (2007) Hydrogen-bonded nucleic acid base pairs containing unusual base tautomers: complete basis set calculations at the MP2 and CCSD (T) levels. J Phys Chem B 111(3):641–645
Koag M-C, Kou Y, Ouzon-Shubeita H, Lee S (2014) Transition-state destabilization reveals how human DNA polymerase β proceeds across the chemically unstable lesion N7-methylguanine. Nucleic Acids Res 42(13):8755–8766
Acknowledgments
The authors acknowledge the general computational facility of the Department of Physics, Assam University, Silchar.
Author information
Authors and Affiliations
Contributions
SB performed simulation, data collection and partial contribution to the first draft of the manuscript. PKS planned and supervised the research work, analysis of the results and writing of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies involving animals performed by any of the authors.
Consent to participate
This article does not contain any studies involving animals performed by any of the authors.
Consent for publication
All the authors mentioned in the manuscript have given consent for submission and subsequent publication of the manuscript.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 162 kb).
Rights and permissions
About this article
Cite this article
Biswas, S., Shukla, P.K. Effect of N7-methylation on base pairing patterns of guanine: a DFT study. J Mol Model 27, 184 (2021). https://doi.org/10.1007/s00894-021-04792-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-021-04792-3