Abstract
A new mycovirus was identified in Trichoderma harzianum strain 137 isolated in Xinjiang province, China. The whole genome sequence of the mycovirus was determined by metagenomic sequencing, RT-PCR, and RACE cloning. The mycovirus contained two genomic segments. The first segment was 2088 bp long and contained a single ORF (ORF1) encoding the RNA-dependent RNA polymerase (RdRP) (72.26 kDa). The second segment was 1634 bp long and also contained a single ORF (ORF2) encoding a hypothetical protein of 37.472 kDa. We named this novel mycovirus “Trichoderma harzianum bipartite mycovirus 1” (ThBMV1). Phylogenetic analysis showed that ThBMV1 clusters with other unclassified dsRNA mycoviruses.
Similar content being viewed by others
Introduction
At present, only three mycoviruses infecting Trichoderma have been described [1,2,3]. In 2009, Jom-in and Akarapisan provided the first description of two mycoviruses, with sizes of 0.7 kb and 1.1 kb, respectively, isolated from Trichoderma [1]. Later, Yun et al. suggested that mycoviruses from Lentinula edodes were widespread in Korea and isolated 32 different dsRNA-containing viruses from 315 strains of Trichoderma spp. [2]. More recently, Lee et al. isolated an unclassified mycovirus from Trichoderma atroviride, naming it “Trichoderma atroviride mycovirus 1” (TaMV 1) [3]. To expand the list of mycoviruses infecting Trichoderma spp., we have isolated 152 Trichoderma spp. from soil samples obtained from the Chinese provinces of Xinjiang, Inner Mongolia, Heilongjiang, and Jilin. These isolates were classified at the species level based on morphological properties and molecular data (sequencing of the ITS region, the translation elongation factor 1-α (tef1-α) gene, and the RNA polymerase subunit II (rpb2) gene). In an effort to identify new Trichoderma viruses, all of these isolates were screened for the presence of mycoviruses (Supplemental Table 1).
Identification of a novel Trichoderma mycovirus
After extraction of dsRNA from mycelia [4], it was possible to visualize two nucleic acid segments with 2 and 1.5 kb, respectively, in strain 137. After extraction, the dsRNA was treated with DNase I and S1 nuclease to confirm the nature of the nucleic acid (Fig. 1A and B). dsRNAs were sent to Shanghai Biotechnology Corporation for sequencing using Illumina HiSeq2500 equipment. Raw data were analyzed by the company, including data preprocessing, sequence assembly, database annotation, gene quantification, species classification, and species abundance analysis. The resulting contigs were annotated using RefSeq by comparing them with the NCBI non-redundant (NR) protein database using BLASTx (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Two contigs of about 2 and 1.5 kb were identified, which are hereafter referred to as RNA1 and RNA2, respectively. Using the EMBOSS-NEEDLE program (https://www.ebi.ac.uk/Tools/emboss/), we found that RNA 1 showed the highest nucleotide sequence similarity (65.7% identity) to Cryphonectria parasitica bipartite mycovirus 1, while RNA 2 showed the highest nucleotide sequence similarity (55.5% identity) to Penicillium aurantiogriseum bipartite virus 1 (Supplemental Table 2). Based on the contig sequences, RT-PCR primers specific for dsRNA1 (forward, AATCGCGCTTACCCGTAACC; reverse, AGTAACTTCGGCACCATCGTCC) and dsRNA2 (forward, AACCACCTCCTCAATCCCTTCC; reverse, TGGGTGACAGTTTTGAAGAGTTGCAGCG) were designed. The RT-PCR assays were repeated to confirm the presence of the virus, and the resulting fragments were sequenced. The resulting sequences consisted of contigs of 1116 and 359 nt (Supplemental Fig. 2). The 5′ and 3′ ends of the bipartite genome were determined using classical RACE methods [5, 6]. The complete genome sequence was assembled using DNAMAN software and deposited in the GenBank database under the accession numbers MH536648 and MH536649.
Detection of dsRNA from Trichoderma harzianum strain 137 by digestion with DNase Ι and S1 nuclease, successively. (A) The dsRNA sample was treated with DNase Ι and electrophoresed in a 1.5% agarose gel and detected on a UV transilluminator. (B) dsRNA sample was treated with S1 nuclease and electrophoresed in a 1.5% agarose gel and detected on a UV transilluminator
Genomic properties
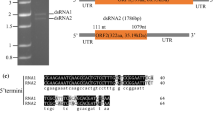
The larger segment of 2088 bp was named dsRNA1, and the smaller segment of 1634 bp was named dsRNA2. Both included a poly(A) tail. The G+C content of the dsRNA1 segment was 50.0%, and for dsRNA2 it was 50.3%. Possible ORFs were found using the NCBI ORF finder tool (https://www.ncbi.nlm.nih.gov/orffinder/). Each dsRNAs contained a single ORF in the positive-sense strand. The 5′ UTR of dsRNA1 was 78 bp long, and the 3′ UTR was 114 bp long; they had no hairpin structures in the UTR regions. The 5′ UTR of dsRNA2 was 105 bp long, while the 3′ UTR was 584 bp long; the structure of UTR regions was the same as for dsRNA1. The 5′ UTRs of dsRNA1 and dsRNA2 had the same conserved element (CUGAGUUAACAAGCCACUGUUUUACUCUCGU), which is necessary for virus replication. A BLASTX search for homologues of ORF1 (nt 79-1974) showed that ORF1 most likely encodes an RNA-dependent RNA polymerase (RdRP) of 631 amino acids with a molecular weight of 72.3 kDa. The C-terminal domain and catalytic palm subdomain were detected in the regions encompassing amino acids 300-448 and 326-454, respectively, using InterPro (http://www.ebi.ac.uk/interpro). Likewise, ORF2 (nt 106-1050) was predicted to encode a hypothetical protein of 314 amino acids with a molecular weight of 37.5 kDa with similarity to other proteins of unknown function or coat proteins from unclassified dsRNA mycoviruses and members of the family Partitiviridae (Supplementary Table 3).
Phylogenetic trees were constructed using the amino acid sequences of RdRps from 29 fungal viruses and hypothetical proteins or coat proteins from 22 mycoviruses. The protein sequences for these viruses were retrieved from the GenBank database and included four bipartite mycoviruses: Cryphonectria parasitica bipartite mycovirus 1, Penicillium aurantiogriseum bipartite virus 1 [7], Heterobasidion RNA virus 6 [8], and Curvularia thermal tolerance virus [9]. Maximum-likelihood phylogenetic trees were constructed using MEGA 7.0 software [10] (Supplemental Fig. 3 and Supplemental Tables 4, 5 and 6) and using the best amino acid substitution model (LG + G for the RdRP and WAG + G + I for the hypothetical or coat protein), and their statistical significance was evaluated by bootstrap (based on 1000 pseudoreplicates). In both phylogenies, the novel mycovirus clustered with a group of unclassified mycoviruses (Fig. 2A and B; Supplemental Tables 4, 5 and 6). For the sake of completeness, an alignment resulting from concatenating the aligned RdRP and hypothetical protein sequences was also used to construct a single maximum-likelihood phylogenetic tree using the WAG+G+I model (Supplemental Fig. 2). The results were identical to those obtained using each protein independently. Based on the above results, we concluded that the mycovirus isolated from T. harzianum isolate 137 was a novel unclassified mycovirus. We have tentatively named it “Trichoderma harzianum bipartite mycovirus 1” (ThBMV1) (Fig. 3).
Schematic representation of the genomic organization of ThBMV1. The dsRNA1 genome contains one ORF. ORF 1 encodes the RNA-dependent RNA polymerase (RdRp). In the RdRp, the C-terminal domain and catalytic domain are located in the regions encompassing amino acids 300-448 and 326-454, respectively. The dsRNA2 genome contains one ORF, ORF 2, which encodes a hypothetical protein. The black lines indicate 5′ and 3′ UTRs
Maximum-likelihood phylogenetic trees inferred from the amino acid sequences of ORF1 and ORF2. Numbers at the nodes represent the statistical support for each cluster (based on 1000 bootstrap replicates). (A) Phylogenetic tree for ORF1 (encoding RdRP), using LG+G+I as the best model of amino acid substitution. (B) Phylogenetic tree for ORF2 (encoding the hypothetical protein), using WAG+G+I as the best model of amino acid substitution
References
Jom-in S, Akarapisan A (2009) Characterization of double-stranded RNA in Trichoderma spp. isolates in Chiang Mai province. J Agric Technol 5:261–270
Yun SH, Song HL, So KK, Kim JM, Kim DH (2016) Incidence of diverse ds RNA mycoviruses in Trichoderma spp. causing green mold disease of shiitake Lentinula edodes. FEMS Microbiol Lett 363:fnw220
Lee S, Yun S, Chun J, Kim D (2017) Characterization of novel dsRNA mycovirus of Trichoderma atroviride NFCF028. Arch Virol 162:1073–1077
Valverde RA, Nameth ST, Jordan RL (1990) Analysis of double-stranded RNA for plant virus diagnosis. Plant Dis 74:255–258
Potgieter AC, Page NA, Liebenberg J, Wright IM, Landt O, van Dijk AA (2009) Improved strategies for sequence-independent amplification and sequencing of viral dsRNA genomes. J Gen Virol 90:1423–1432
Hou Z, Xue C, Peng Y, Katan T, Kistler HC, Xu JR (2002) A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol Plant Microbe Interact 15:1119–1127
Nerva L, Ciuffo M, Vallino M, Margaria P, Varese GC, Gnavi G, Turina M (2016) Multiple approaches for the detection and characterization of viral and plasmid symbionts from a collection of marine fungi. Virus Res 19:22–38
Kashif M, Hyder R, De Vega Perez D, Hantula J, Vainio EJ (2015) Heterobasidion wood decay fungi host diverse and globally distributed viruses related to Helicobasidium mompa partitivirus V70. Virus Res 195:119–123
Márquez LM, Redman RS, Rodriguez RJ, Roossinck MJ (2007) A virus in a fungus in a plant: three-way symbiosis required for thermal tolerance. Science 215:513–515
Tamura K, Da Peterson, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Acknowledgements
We thank Irina S. Druzhinina (Institute of Chemical, Environmental and Bioscience Engineering, Department of Biochemical Technology, Microbiology and Applied Genomic Group, TU WIEN, Gumpendorferstrasse 1a, A1060, Vienna, Austria) for her insights and suggestions for the identification of the Trichoderma strains.
Funding
This work was supported by the National Key Research and Development Plan (Chemical fertilizer and pesticide reducing efficiency synergistic technology research and development): Research and demonstration of a new high efficiency biocide (2017YFD0201100-2017YFD0201102) granted to Xiliang Jiang; Research and demonstration of technology integration on reducing chemical fertilizer and pesticide application in open field vegetables (2018YFD0201200-2018YFD0201202) granted to Mei Li; Demonstration of comprehensive prevention and control technology of non-point source pollution in main vegetable producing areas of Huang Huai Hai (SQ2018YFD080026) granted to Beilei Wu; Work in València was supported by Spain’s Agencia Estatal de Investigación - FEDER grant BFU2015-65037-P granted to Santiago F. Elena.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
None of the authors have conflicts of interest.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Informed consent
Not applicable.
Additional information
Handling Editor: Robert H. A. Coutts.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
OpenAccess This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Liu, C., Li, M., Redda, E.T. et al. Complete nucleotide sequence of a novel mycovirus from Trichoderma harzianum in China. Arch Virol 164, 1213–1216 (2019). https://doi.org/10.1007/s00705-019-04145-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-019-04145-9