Abstract
The complete genome sequence of a Capsicum chlorosis virus from China (CaCV-Hainan) was determined. The tripartite genome of CaCV-Hainan consists of small (S), medium (M), and large (L) RNAs of 3629, 4859, and 8912 nucleotides (nt), respectively. The S and M RNAs contain intergenic regions (IGRs) of 1348 and 462 nt, respectively. Strikingly, sequence comparisons among CaCV isolates revealed that the S RNA IGR of CaCV-Hainan derived from the CaCV-Qld-3432 Australia isolate through deletion of two stretches of 25- and 325-nt sequences within the S RNA IGR of CaCV-Qld-3432. Moreover, the S RNA IGR of CaCV-Hainan was inserted with two stretches of 10- and 20-nt sequences of an unknown origin. The S RNA IGR of CaCV-Ph from Taiwan and CaCV-NRA from Thailand also derived from the CaCV-Qld-3432 through deletion of 218-nt sequences. Our findings provide valuable new insight into the structural variations and evolutionary origin of CaCV IGRs.

Similar content being viewed by others
References
Plyusnin A, Beaty BJ, Elliott RM, Goldbach R, Kormelink R, Lundkvist Å, Schmaljohn CS, Tesh RB (2012) Bunyaviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, pp 725–741
Chen CC, Huang CH, Cheng YH, Chen TC, Yeh SD, Chang CA (2009) First report of Capsicum chlorosis virus infecting amaryllis and blood lily in Taiwan. Plant Dis 93:1346
Chen K, Xu Z, Yan L, Wang G (2007) Characterization of a new strain of Capsicum chlorosis virus from peanut (Arachis hypogaea L.) in China. J Phytopathol 155:178–181
Chen TC, Li JT, Lin YP, Yeh YC, Kang YC, Huang LH, Yeh SD (2012) Genomic characterization of Calla lily chlorotic spot virus and design of broad-spectrum primers for detection of tospoviruses. Plant Pathol 61:183–194
Chiemsombat P, Gajanandana O, Warin N, Hongprayoon R, Bhunchoth A, Pongsapich P (2008) Biological and molecular characterization of tospoviruses in Thailand. Arch Virol 153:571–577
Hanssen IM, Lapidot M, Thomma BP (2010) Emerging viral diseases of tomato crops. Mol Plant Microbe Interact 23:539–548
Hassani-Mehraban A, Saaijer J, Peters D, Goldbach R, Kormelink R (2007) Molecular and biological comparison of two Tomato yellow ring virus (TYRV) isolates: challenging the Tospovirus species concept. Arch Virol 152:85–96
Knierim D, Blawid R, Maiss E (2006) The complete nucleotide sequence of a capsicum chlorosis virus isolate from Lycopersicum escultentum in Thailand. Arch Virol 151:1761–1782
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Kunkalikar SR, Sudarsana P, Rajagopalan P, Zehr UB, Ravi KS (2010) Biological and molecular characterization of Capsicum chlorosis virus infecting chilli and tomato in India. Arch Virol 155:1047–1057
McMichael LA, Persley DM, Thomas JE (2002) A new tospovirus serogroup IV species infecting capsicum and tomato in Queensland, Australia. Aust Plant Pathol 31:231–239
Melzer MJ, Shimabukuro J, Long MH, Nelson SC, Alvarez AM, Borth WB, Hu JS (2014) First report of Capsicum chlorosis virus infecting waxflower (Hoya calycina Schlecter) in the United States. Plant Dis 98:571
Pappu HR, Jones RA, Jain RK (2009) Global status of tospovirus epidemics in diverse cropping systems: successes achieved and challenges ahead. Virus Res 141:219–236
Pappu S, Bhat A, Pappu H, Deom C, Culbreath A (2000) Phylogenetic studies of tospoviruses (family: Bunyaviridae) based on intergenic region sequences of small and medium genomic RNAs. Arch Virol 145:1035–1045
Pappu S, Bhat A, Pappu H, Deom C, Culbreath A (2000) Phylogenetic studies of tospoviruses (family: Bunyaviridae) based on intergenic region sequences of small and medium genomic RNAs. Arch Virol 145:1035–1045
Persley D, Thomas J, Sharman M (2006) Tospoviruses—an Australian perspective. Aust Plant Pathol 35:161–180
Prins M, Goldbach R (1998) The emerging problem of tospovirus infection and nonconventional methods of control. Trends Microbiol 6:31–35
Rao X, Li Y, Wu Z, Liu Y, Wang W (2014) Expression of nucleocapsid protein of capsicum chlorosis virus in a prokaryotic system and its application in virus detection. J Plant Pathol 96:381–384
Turina M, Tavella L, Ciuffo M (2012) Tospoviruses in the mediterranean area. Adv Virus Res 84:403–437
Widana Gamage S, Persley DM, Higgins CM, Dietzgen RG (2015) First complete genome sequence of a Capsicum chlorosis tospovirus isolate from Australia with an unusually large S RNA intergenic region. Arch Virol 160:869–872
Yin YY, Li TT, Lu X, Fang Q, Ding M, Zhang ZK (2015) First report of Capsicum chlorosis virus infecting tomato in Yunnan, southwest of China. Plant Dis 100:230
Zheng YX, Chen CC, Jan FJ (2011) Complete nucleotide sequence of capsicum chlorosis virus isolated from Phalaenopsis orchid and the prediction of the unexplored genetic information of tospoviruses. Arch Virol 156:421–432
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (31630062 and 31471746), the Special Fund for Agro-Scientific Research in the Public Interest (201303028), the Youth Talent Support Program of China and Distinguished Professor of Jiangsu Province to X. T., the Fundamental Research Funds for the Central Universities (KYTZ201403), the Specialized Research Fund for the Doctoral Program of Higher Education (20130097110004).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
705_2017_3448_MOESM2_ESM.pdf
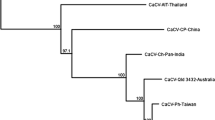
Supplementary material 2 Supplemental Figure 1: Alignment of nucleotide sequences of S RNA intergenic regions (IGRs) among CaCV-Qld-3432, CaCV-Hainan, CaCV-Ph, CaCV-NRA, CaCV-Ch-Pan, and CaCV-AIT. The nucleotide sites that are identical among the CaCV isolates are highlighted. The accession numbers of the IGR sequences of the CaCV isolates, and of watermelon silver mottle virus (WSMoV) and tomato zonate spot virus (TZSV), are provided in the main text. Supplemental Figure 2: Phylogenetic analysis of the nucleotide sequences of S RNA intergenic regions (IGRs) of CaCV-Qld-3432, CaCV-Hainan, CaCV-Ph, CaCV-NRA, CaCV-Ch-Pan, and CaCV-AIT. The phylogenetic tree was constructed using the maximum likelyhood method. Bootstrap values on the branches are derived from a 1,000-bootstrap replicate analysis. The accession numbers of the IGR sequences of the CaCV isolates are described in the main text. The scale bar represents the genetic distance (PDF 80 kb)
Rights and permissions
About this article
Cite this article
Huang, Y., Hong, H., Zhao, XH. et al. Complete genome sequence of a Capsicum chlorosis virus in China and the structural variation and evolutionary origin of its S RNA intergenic region. Arch Virol 162, 3229–3232 (2017). https://doi.org/10.1007/s00705-017-3448-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-017-3448-4