Abstract
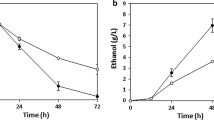
Industrial production of lignocellulosic ethanol requires a microorganism utilizing both hexose and pentose, and tolerating inhibitors. In this study, a hydrolysate-cofermenting Saccharomyces cerevisiae strain was obtained through one step in vivo DNA assembly of pentose-metabolizing pathway genes, followed by consecutive adaptive evolution in pentose media containing acetic acid, and direct screening in biomass hydrolysate media. The strain was able to coferment glucose and xylose in synthetic media with the respective maximal specific rates of glucose and xylose consumption, and ethanol production of 3.47, 0.38 and 1.62 g/g DW/h, with an ethanol titre of 41.07 g/L and yield of 0.42 g/g. Industrial wheat straw hydrolysate fermentation resulted in maximal specific rates of glucose and xylose consumption, and ethanol production of 2.61, 0.54 and 1.38 g/g DW/h, respectively, with an ethanol titre of 54.11 g/L and yield of 0.44 g/g. These are among the best for wheat straw hydrolysate fermentation through separate hydrolysis and cofermentation.






Similar content being viewed by others
References
Geng AL (2013) Conversion of oil palm empty fruit bunch to biofuels. In: Fang Z (ed) Biofuels, book 3. INTECH Open Access Publisher, Shanghai (ISBN:9535110500)
Kwak S, Jin YS (2017) Production of fuels and chemicals from xylose by engineered Saccharomyces cerevisiae: a review and perspective. Microb Cell Fact 16:82
Tran Nguyen Hoang P, Ko JK, Gong G, Um Y, Lee SM (2018) Genomic and phenotypic characterization of a refactored xylose-utilizing Saccharomyces cerevisiae strain for lignocellulosic biofuel production. Biotechnol Biofuels 11:268
Wang C, Zhao J, Qiu C, Wang S, Shen Y, Du B, Ding Y, Bao X (2017) Coutilization of d-glucose, d-xylose, and l-arabinose in Saccharomyces cerevisiae by coexpressing the metabolic pathways and evolutionary engineering. Biomed Res Int 2017:5318232
Garcia SR, Karhumaa K, Fonseca C, Sanchez NV, Almeida JR, Larsson CU, Bengtsson O, Bettiga M, Hahn-Hagerdal B, Gorwa-Grauslund MF (2010) Improved xylose and arabinose utilization by an industrial recombinant Saccharomyces cerevisiae strain using evolutionary engineering. Biotechnol Biofuels 3:13
Hahn-Hägerdal B, Karhumaa K, Fonseca C, Spencer-Martins I, Gorwa-Grauslund MF (2007) Towards industrial pentose-fermenting yeast strains. Appl Microbiol Biotechnol 74:937–953
Peng B, Shen Y, Li X, Chen X, Hou J, Bao X (2012) Improvement of xylose fermentation in respiratory-deficient xylose-fermenting Saccharomyces cerevisiae. Metab Eng 14:9–18
Peng B, Huang S, Liu T, Geng A (2015) Bacterial xylose isomerases from the mammal gut Bacteroidetes cluster function in Saccharomyces cerevisiae for effective xylose fermentation. Microb Cell Fact 14:70
Bettiga M, Hahn-Hägerdal B, Gorwa-Grauslund MF (2008) Comparing the xylose reductase/xylitol dehydrogenase and xylose isomerase pathways in arabinose and xylose fermenting Saccharomyces cerevisiae strains. Biotechnol Biofuels 1:16
Jo JH, Park YC, Jin YS, Seo JH (2017) Construction of efficient xylose-fermenting Saccharomyces cerevisiae through a synthetic isozyme system of xylose reductase from Scheffersomyces stipitis. Bioresour Technol 241:88–94
Eliasson A, Christensson C, Wahlbom CF, Hahn-Hägerdal B (2000) Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl Environ Microbiol 66:3381–3386
Wisselink HW, Toirkens MJ, Del Rosario Franco Berriel M, Winkler AA, van Dijken JP, Pronk JT, van Maris AJA (2007) Engineering of Saccharomyces cerevisiae for efficient anaerobic alcoholic fermentation of l-arabinose. Appl Environ Microbiol 73:4881–4891
Wisselink HW, Toirkens MJ, Wu Q, Pronk JT, van Maris AJA (2009) Novel evolutionary engineering approach for accelerated utilization of glucose, xylose, and arabinose mixtures by engineered Saccharomyces cerevisiae strains. Appl Environ Microbiol 75:907–914
Chandel AK, da Silva SS, Singh OV (2011) Detoxification of lignocellulosic hydrolysates for improved bioethanol production. In: dos Santos Bernardes MA (ed) Biofuel production—recent developments and prospects. INTECH Open Access Publisher, Shanghai
Koppram R, Albers E, Olsson L (2012) Evolutionary engineering strategies to enhance tolerance of xylose utilizing recombinant yeast to inhibitors derived from spruce biomass. Biotechnol Biofuels 5:32
Li H, Shen Y, Wu M, Hou J, Jiao C, Li Z, Liu X, Bao X (2016) Engineering a wild-type diploid Saccharomyces cerevisiae strain for second-generation bioethanol production. Bioresour Bioprocess 3:51
Demeke MM, Dietz H, Li Y, Foulquie-Moreno MR, Mutturi S, Deprez S, Den Abt T, Bonini BM, Liden G, Dumortier F et al (2013) Development of a d-xylose fermenting and inhibitor tolerant industrial Saccharomyces cerevisiae strain with high performance in lignocellulose hydrolysates using metabolic and evolutionary engineering. Biotechnol Biofuels 6:89
Chen Y, Stabryla L, Wei N (2016) Improved acetic acid resistance in Saccharomyces cerevisiae by overexpression of the WHI2 gene identified through inverse metabolic engineering. Appl Environ Microbiol 82:2156–2166
Guadalupe-Medina V, Metz B, Oud B, van Der Graaf CM, Mans R, Pronk JT, van Maris AJA (2014) Evolutionary engineering of a glycerol-3-phosphate dehydrogenase-negative, acetate-reducing Saccharomyces cerevisiae strain enables anaerobic growth at high glucose concentrations. Microb Biotechnol 7:44–53
Hahn-Hägerdal B, Karhumaa K, Jeppsson M, Gorwa-Grauslund MF (2007) Metabolic engineering for pentose utilization in Saccharomyces cerevisiae. Adv Biochem Eng Biotechnol 108:147–177
Shao Z, Zhao H, Zhao H (2009) DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res DNA 37:e16
Shen FL, Huang SC, Hou PC, Geng AL, Ruan WQ (2017) A high effective autonomous replicative sequence in Saccharomyces cerevisiae. Food Ferment Ind 3:20–25
Jorgensen H (2009) Effect of nutrients on fermentation of pretreated wheat straw at very high dry matter content by Saccharomyces cerevisiae. Appl Biochem Biotechnol 153:44–57
Wang Z, Ong HX, Geng A (2012) Cellulase production and oil palm empty fruit bunch saccharification by a new isolate of Trichoderma koningii D-64. Proc Biochem 47:1564–1571
Güldener U, Heck S, Fiedler T, Beinhauer J, Hegemann JH (1966) A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res 24:2519–2524
Yanisch-Perron C, Vieira J, Messing J (1985) Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119
Gietz RD, Schiestl RH (2007) Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2:38–41
Kim B, Du J, Eriksen DT, Zhao H (2013) Combinatorial design of a highly efficient xylose-utilizing pathway in Saccharomyces cerevisiae for the production of cellulosic biofuels. Appl Environ Microb 79:931–941
Sedlak M, Ho NW (2004) Production of ethanol from cellulosic biomass hydrolysates using genetically engineered Saccharomyces yeast capable of cofermenting glucose and xylose. Appl Biochem Biotechnol 116:403–416
Latimer LN, Lee ME, Medina-Cleghorn D, Kohnz RA, Nomura DK, Dueber JE (2014) Employing a combinatorial expression approach to characterize xylose utilization in Saccharomyces cerevisiae. Metab Eng 25:20–29
Zhou H, Cheng JS, Wang BL, Fink GR, Stephanopoulos G (2012) Xylose isomerase overexpression along with engineering of the pentose phosphate pathway and evolutionary engineering enable rapid xylose utilization and ethanol production by Saccharomyces cerevisiae. Metab Eng 14:611–622
Liu TT, Huang SC, Geng AL (2018) Recombinant diploid Saccharomyces cerevisiae strain development for rapid glucose and xylose co-fermentation. Fermentation 4:59
Klimacek M, Kirl E, Krahulec S, Longus K, Novy V, Nidetzky B (2014) Stepwise metabolic adaption from pure metabolization to balanced anaerobic growth on xylose explored for recombinant Saccharomyces cerevisiae. Microb Cell Fact 13:37
Li YC, Mitsumasu K, Gou ZX, Gou M, Tang YQ, Li GY, Wu XL, Akamatsu T, Taguchi H, Kida K (2016) Xylose fermentation efficiency and inhibitor tolerance of the recombinant industrial Saccharomyces cerevisiae strain NAPX37. Appl Microbiol Biotechnol 100:1531–1542
Novy V, Wang R, Westman JO, Franzén CJ, Nidetzky B (2017) Saccharomyces cerevisiae strain comparison in glucose-xylose fermentations on defined substrates and in high-gravity SSCF: convergence in strain performance despite differences in genetic and evolutionary engineering history. Biotechnol Biofuels 10:205
Papapetridis I, Verhoeven MD, Wiersma SJ, Goudriaan M, van Maris AJA, Pronk JT (2018) Laboratory evolution for forced glucose-xylose co-consumption enables identification of mutations that improve mixed-sugar fermentation by xylose-fermenting Saccharomyces cerevisiae. FEMS Yeast Res 18(6):foy056
Lee YG, Jin YS, Cha YL, Seo JH (2017) Bioethanol production from cellulosic hydrolysates by engineered industrial Saccharomyces cerevisiae. Bioresour Technol 228:355–361
Yuan Z, Li G, Hegg EL (2018) Enhancement of sugar recovery and ethanol production from wheat straw through alkaline pre-extraction followed by steam pretreatment. Bioresour Technol 266:194–202
Wallace-Salinas V, Gorwa-Grauslund MF (2013) Adaptive evolution of an industrial strain of Saccharomyces cerevisiae for combined tolerance to inhibitors and temperature. Biotechnol Biofuels 6:151
Smith J, van Rensburg E, Görgens JF (2014) Simultaneously improving xylose fermentation and tolerance to lignocellulosic inhibitors through evolutionary engineering of recombinant Saccharomyces cerevisiae harbouring xylose isomerase. BMC Biotechnol 14:41
Kim SK, Jin YS, Choi IG, Park YC, Seo JH (2015) Enhanced tolerance of Saccharomyces cerevisiae to multiple lignocellulose-derived inhibitors through modulation of spermidine contents. Metab Eng 29:46–55
Li YC, Gou ZX, Zhang Y, Xia ZY, Tang YQ, Kida K (2017) Inhibitor tolerance of a recombinant flocculating industrial Saccharomyces cerevisiae strain during glucose and xylose co-fermentation. Braz J Microbiol 48:791–800
Jung YH, Kim IJ, Han JI, Choi IG, Kim KH (2011) Aqueous ammonia pretreatment of oil palm empty fruit bunches for ethanol production. Bioresour Technol 102:9806–9809
Jung YH, Kim IJ, Kim HK, Kim KH (2013) Dilute acid pretreatment of lignocellulose for whole slurry ethanol fermentation. Bioresour Technol 132:109–114
Cui X, Zhao X, Zeng J, Loh SK, Choo YM, Liu D (2014) Robust enzymatic hydrolysis of Formiline-pretreated oil palm empty fruit bunches (EFB) for efficient conversion of polysaccharide to sugars and ethanol. Bioresour Technol 166:584–591
Duangwang S, Ruengpeerakul T, Cheirsilp B, Yamsaengsung R, Sangwichien C (2016) Pilot-scale steam explosion for xylose production from oil palm empty fruit bunches and the use of xylose for ethanol production. Bioresour Technol 203:252–258
Qiu J, Ma L, Shen F, Yang G, Zhang Y, Deng S, Zhang J, Zeng Y, Hu Y (2017) Pretreating wheat straw by phosphoric acid plus hydrogen peroxide for enzymatic saccharification and ethanol production at high solid loading. Bioresour Technol 238:174–181
Qiu J, Tian D, Shen F, Hu J, Zeng Y, Yang G, Zhang Y, Deng S, Zhang J (2018) Bioethanol production from wheat straw by phosphoric acid plus hydrogen peroxide (PHP) pretreatment via simultaneous saccharification and fermentation (SSF) at high solid loadings. Bioresour Technol 268:355–362
Mahboubi A, Ylitervo P, Doyen W, De Wever H, Molenberghs B, Taherzadeh MJ (2017) Continuous bioethanol fermentation from wheat straw hydrolysate with high suspended solid content using an immersed flat sheet membrane bioreactor. Bioresour Technol 241:296–308
Cassells B, Karhumaa K, Sànchez I, Nogué V, Lidén G (2017) Hybrid SSF/SHF processing of SO2 pretreated wheat straw-tuning co-fermentation by yeast inoculum size and hydrolysis time. Appl Biochem Biotechnol 181:536–547
Westman JO, Wang R, Novy V, Franzén CJ (2017) Sustaining fermentation in high-gravity ethanol production by feeding yeast to a temperature-profiled multifeed simultaneous saccharification and co-fermentation of wheat straw. Biotechnol Biofuels 10:213
Acknowledgements
This study was funded by the Science and Engineering Research Council of the Agency for Science Technology and Research (A*STAR) Singapore (Grant no. 092 139 0035). The authors are grateful for the industrial lignocellulose hydrolysate samples provided by Teck Guan Holdings Sdn Bhd, Tawau, Malaysia and Inbicon A/S, Fredericia, Denmark. We are also thankful for the internship opportunities provided by Ngee Ann Polytechnic Singapore to Shuangcheng Huang and Tingting Liu.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Huang, S., Liu, T., Peng, B. et al. Enhanced ethanol production from industrial lignocellulose hydrolysates by a hydrolysate-cofermenting Saccharomyces cerevisiae strain. Bioprocess Biosyst Eng 42, 883–896 (2019). https://doi.org/10.1007/s00449-019-02090-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-019-02090-0