Abstract
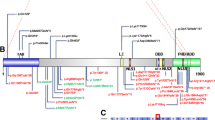
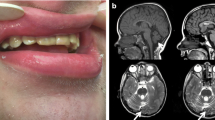
CHD8, which encodes Chromodomain helicase DNA-binding protein 8, is one of a few well-established Autism Spectrum Disorder (ASD) genes. Over 60 mutations have been reported in subjects with variable phenotypes, but little is known concerning genotype–phenotype correlations. We have identified four novel de novo mutations in Chinese subjects: two nonsense variants (c.3562C>T/p.Arg1188X, c.2065C>A/p.Glu689X), a splice site variant (c.4818-1G>A) and a missense variant (c.3502T>A/p.Tyr1168Asn). Three of these were identified from a 445-member ASD cohort by ASD gene panel sequencing of the 96 subjects who remained negative after molecular testing for copy number variation, Rett syndrome, FragileX and tuberous sclerosis complex (TSC). The fourth (p.Glu689X) was detected separately by diagnostic trio exome sequencing. We used diagnostic instruments and a comprehensive review of phenotypes, including prenatal and postnatal growth parameters, developmental milestones, and dysmorphic features to compare these four subjects. In addition to autism, they also presented with prenatal onset macrocephaly, intellectual disability, overgrowth during puberty, sleep disorder, and dysmorphic features, including broad forehead with prominent supraorbital ridges, flat nasal bridge, telecanthus and large ears. For further comparison, we compiled a comprehensive list of CHD8 variants from the literature and databases, which revealed constitutive and somatic truncating variants in the HELIC (Helicase-C) domain in ASD and in cancer patients, respectively, but not in the general population. Furthermore, HELIC domain mutations were associated with a severe phenotype defined by a greater number of clinical features, lower verbal IQ, and a prominent, consistent pattern of overgrowth as measured by weight, height and head circumference. Overall, this study adds to the ASD-associated loss-of-function mutations in CHD8 and highlights the clinical importance of the HELIC domain of CHD8.





Similar content being viewed by others
Data availability
gnomAD, gnomad.broadinstitute.org/;ExAC Browse, https://Exac.broadinstitute.org/; Mutation Taster, https://mutationtaster.org/, Polyphen-2, https://genetics.bwh.harvard.edu/pph2/; SIFT, https://sif.jcvi.org, UCSC human Genome Browser, https://genome-euro.ucsc.edu/index.html; Exome Variant Server, https://evs.gs.washington.edu/EVS/;1K Genomes Project: https://www.1000genomes.org/; BioMuta v2.0 (https://hive.biochemistry.gwu.edu/cgi-bin/prd/biomuta/servlet.cgi); Conserved Domain Database (CDD) https://www.ncbi.nlm.nih.gov/Structure/cdd/.
References
Anzo M, Takahashi T, Sato S, Matsuo N (2002) The cross-sectional head circumference growth curves for Japanese from birth to 18 years of age: the 1990 and 1992–1994 national survey data. Ann Hum Biol 29:373–388. https://doi.org/10.1080/03014460110089526
Bernardo P, Galletta D, Iasevoli F, D'Ambrosio L, Troisi S, Gennaro E, Zara F, Striano S, de Bartolomeis A, Coppola A (2017) CHD2 mutations: only epilepsy? Description of cognitive and behavioral profile in a case with a new mutation. Seizure-Eur J Epilepsy 51:186–189. https://doi.org/10.1016/j.seizure.2017.09.001
Bernier R, Golzio C, Xiong B, Stessman HA, Coe BP, Penn O, Witherspoon K, Gerdts J, Baker C, Vulto-van Silfhout AT, Schuurs-Hoeijmakers JH, Fichera M, Bosco P, Buono S, Alberti A, Failla P, Peeters H, Steyaert J, Vissers LE, Francescatto L, Mefford HC, Rosenfeld JA, Bakken T, O'Roak BJ, Pawlus M, Moon R, Shendure J, Amaral DG, Lein E, Rankin J, Romano C, de Vries BB, Katsanis N, Eichler EE (2014) Disruptive CHD8 mutations define a subtype of autism early in development. Cell 158:263–276. https://doi.org/10.1016/j.cell.2014.06.017
Blok LS, Rousseau J, Twist J, Ehresmann S, Takaku M, Venselaar H, Rodan LH, Nowak CB, Douglas J, Swoboda KJ, Steeves MA, Sahai I, Stumpel CTRM, Stegmann APA, Wheeler P, Willing M, Fiala E, Kochhar A, Gibson WT, Cohen ASA, Agbahovbe R, Innes AM, Au PYB, Rankin J, Anderson IJ, Skinner SA, Louie RJ, Warren HE, Afenjar A, Keren B, Nava C, Buratti J, Isapof A, Rodriguez D, Lewandowski R, Propst J, van Essen T, Choi M, Lee S, Chae JH, Price S, Schnur RE, Douglas G, Wentzensen IM, Zweier C, Reis A, Bialer MG, Moore C, Koopmans M, Brilstra EH, Monroe GR, van Gassen KLI, van Binsbergen E, Newbury-Ecob R, Bownass L, Bader I, Mayr JA, Wortmann SB, Jakielski KJ, Strand EA, Kloth K, Bierhals T, Roberts JD, Petrovich RM, Machida S, Kurumizaka H, Lelieveld S, Pfundt R, Jansen S, Deriziotis P, Faive L, Thevenon J, Assoum M, Shriberg L, Kleefstra T, Brunner HG, Wade PA, Fisher SE, Campeau PM (2018) CHD3 helicase domain mutations cause a neurodevelopmental syndrome with macrocephaly and impaired speech and language. Nat Commun. https://doi.org/10.1038/S41467-018-06014-6
Chang X, Wang K (2012) wANNOVAR: annotating genetic variants for personal genomes via the web. J Med Genet 49:433–436. https://doi.org/10.1136/jmedgenet-2012-100918
De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, Singh T, Klei L, Kosmicki J, Shih-Chen F, Aleksic B, Biscaldi M, Bolton PF, Brownfeld JM, Cai J, Campbell NG, Carracedo A, Chahrour MH, Chiocchetti AG, Coon H, Crawford EL, Curran SR, Dawson G, Duketis E, Fernandez BA, Gallagher L, Geller E, Guter SJ, Hill RS, Ionita-Laza J, Jimenz Gonzalez P, Kilpinen H, Klauck SM, Kolevzon A, Lee I, Lei I, Lei J, Lehtimaki T, Lin CF, Ma'ayan A, Marshall CR, McInnes AL, Neale B, Owen MJ, Ozaki N, Parellada M, Parr JR, Purcell S, Puura K, Rajagopalan D, Rehnstrom K, Reichenberg A, Sabo A, Sachse M, Sanders SJ, Schafer C, Schulte-Ruther M, Skuse D, Stevens C, Szatmari P, Tammimies K, Valladares O, Voran A, Li-San W, Weiss LA, Willsey AJ, Yu TW, Yuen RK, Cook EH, Freitag CM, Gill M, Hultman CM, Lehner T, Palotie A, Schellenberg GD, Sklar P, State MW, Sutcliffe JS, Walsh CA, Scherer SW, Zwick ME, Barett JC, Cutler DJ, Roeder K, Devlin B, Daly MJ, Buxbaum JD (2014) Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515:209–215. https://doi.org/10.1038/nature13772
Dong C, Wei P, Jian X, Gibbs R, Boerwinkle E, Wang K, Liu X (2015) Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet 24:2125–2137. https://doi.org/10.1093/hmg/ddu733
Farnung L, Vos SM, Wigge C, Cramer P (2017) Nucleosome-Chd1 structure and implications for chromatin remodelling. Nature 550:539. https://doi.org/10.1038/nature24046
Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim YC, Minor W, Rastinejad F, Khorasanizadeh S (2005) Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature 438:1181–1185. https://doi.org/10.1038/nature04290
Fukai R, Hiraki Y, Yofune H, Tsurusaki Y, Nakashima M, Saitsu H, Tanaka F, Miyake N, Matsumoto N (2015) A case of autism spectrum disorder arising from a de novo missense mutation in POGZ. J Hum Genet 60:277–279. https://doi.org/10.1038/jhg.2015.13
Gauthier J, Champagne N, Lafreniere RG, Xiong L, Spiegelman D, Brustein E, Lapointe M, Peng H, Cote M, Noreau A, Hamdan FF, Addington AM, Rapoport JL, Delisi LE, Krebs MO, Joober R, Fathalli F, Mouaffak F, Haghighi AP, Neri C, Dube MP, Samuels ME, Marineau C, Stone EA, Awadalla P, Barker PA, Carbonetto S, Drapeau P, Rouleau GA (2010) De novo mutations in the gene encoding the synaptic scaffolding protein SHANK3 in patients ascertained for schizophrenia. Proc Natl Acad Sci USA 107:7863–7868. https://doi.org/10.1073/pnas.0906232107
Geisheker MR, Heymann G, Wang TY, Coe BP, Turner TN, Stessman HAF, Hoekzema K, Kvarnung M, Shaw M, Friend K, Liebelt J, Barnett C, Thompson EM, Haan E, Guo H, Anderlid BM, Nordgren A, Lindstrand A, Vandeweyer G, Alberti A, Avola E, Vinci M, Giusto S, Pramparo T, Pierce K, Nalabolu S, Michaelson JJ, Sedlacek Z, Santen GWE, Peeters H, Hakonarson H, Courchesne E, Romano C, Kooy RF, Bernier RA, Nordenskjold M, Gecz J, Xia K, Zweifel LS, Eichler EE (2017) Hotspots of missense mutation identify neurodevelopmental disorder genes and functional domains. Nat Neurosci 20:1043. https://doi.org/10.1038/nn.4589
Hauk G, McKnight JN, Nodelman IM, Bowman GD (2010) The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol Cell 39:711–723. https://doi.org/10.1016/j.molcel.2010.08.012
International Molecular Genetic Study of Autism Consortium (2001) A genomewide screen for autism: strong evidence for linkage to chromosomes 2q, 7q, and 16p. Am J Hum Genet 69:570–581. https://doi.org/10.1086/323264
Kimura H, Wang C, Ishizuka K, Xing J, Takasaki Y, Kushima I, Aleksic B, Uno Y, Okada T, Ikeda M, Mori D, Inada T, Iwata N, Ozaki N (2016) Identification of a rare variant in CHD8 that contributes to schizophrenia and autism spectrum disorder susceptibility. Schizophr Res 178:104–106. https://doi.org/10.1016/j.schres.2016.08.023
Krumm N, O'Roak BJ, Shendure J, Eichler EE (2014) A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci 37:95–105. https://doi.org/10.1016/j.tins.2013.11.005
Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S (2016) ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. https://doi.org/10.1093/nar/gkv1222
Lee H, Deignan JL, Dorrani N, Strom SP, Kantarci S, Quintero-Rivera F, Das K, Toy T, Harry B, Yourshaw M, Fox M, Fogel BL, Martinez-Agosto JA, Wong DA, Chang VY, Shieh PB, Palmer CG, Dipple KM, Grody WW, Vilain E, Nelson SF (2014) Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA 312:1880–1887. https://doi.org/10.1001/jama.2014.14604
Manning BJ, Yusufzai T (2017) The ATP-dependent chromatin remodeling enzymes CHD6, CHD7, and CHD8 exhibit distinct nucleosome binding and remodeling activities. J Biol Chem. https://doi.org/10.1074/jbc.M117.779470
McCarthy SE, Gillis J, Kramer M, Lihm J, Yoon S, Berstein Y, Mistry M, Pavlidis P, Solomon R, Ghiban E, Antoniou E, Kelleher E, O'Brien C, Donohoe G, Gill M, Morris DW, McCombie WR, Corvin A (2014) De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol Psychiatry 19:652–658. https://doi.org/10.1038/mp.2014.29
Michaelson JJ, Shi Y, Gujral M, Zheng H, Malhotra D, Jin X, Jian M, Liu G, Greer D, Bhandari A, Wu W, Corominas R, Peoples A, Koren A, Gore A, Kang S, Lin GN, Estabillo J, Gadomski T, Singh B, Zhang K, Akshoomoff N, Corsello C, McCarroll S, Iakoucheva LM, Li Y, Wang J, Sebat J (2012) Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell 151:1431–1442. https://doi.org/10.1016/j.cell.2012.11.019
Nellhaus G (1968) Head circumference from birth to eighteen years. Practical composite international and interracial graphs. Pediatrics 41:106–114
Nemirovsky SI, Cordoba M, Zaiat JJ, Completa SP, Vega PA, Gonzalez-Moron D, Medina NM, Fabbro M, Romero S, Brun B, Revale S, Ogara MF, Pecci A, Marti M, Vazquez M, Turjanski A, Kauffman MA (2015) Whole genome sequencing reveals a de novo SHANK3 mutation in familial autism spectrum disorder. PLoS ONE 10:e0116358. https://doi.org/10.1371/journal.pone.0116358
Nodelman IM, Bleichert F, Patel A, Ren R, Horvath KC, Berger JM, Bowman GD (2017) Interdomain communication of the Chd1 chromatin remodeler across the DNA Gyres of the nucleosome. Mol Cell 65(447–459):e6. https://doi.org/10.1016/j.molcel.2016.12.011
O'Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, Carvill G, Kumar A, Lee C, Ankenman K, Munson J, Hiatt JB, Turner EH, Levy R, O'Day DR, Krumm N, Coe BP, Martin BK, Borenstein E, Nickerson DA, Mefford HC, Doherty D, Akey JM, Bernier R, Eichler EE, Shendure J (2012) Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338:1619–1622. https://doi.org/10.1126/science.1227764
O'Roak BJ, Stessman HA, Boyle EA, Witherspoon KT, Martin B, Lee C, Vives L, Baker C, Hiatt JB, Nickerson DA, Bernier R, Shendure J, Eichler EE (2014) Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat Commun 5(1):5595
Pilarowski GO, Vernon HJ, Applegate CD, Boukas L, Cho MT, Gurnett CA, Benke PJ, Beaver E, Heeley JM, Medne L, Krantz ID, Azage M, Niyazov D, Henderson LB, Wentzensen IM, Baskin B, Sacoto MJG, Bowman GD, Bjornsson HT (2018) Missense variants in the chromatin remodeler CHD1 are associated with neurodevelopmental disability. J Med Genet 55:561–566. https://doi.org/10.1136/jmedgenet-2017-104759
Popp B, Ekici AB, Thiel CT, Hoyer J, Wiesener A, Kraus C, Reis A, Zweier C (2017) Exome Pool-Seq in neurodevelopmental disorders. Eur J Hum Genet 25:1364–1376. https://doi.org/10.1038/s41431-017-0022-1
Prontera P, Ottaviani V, Toccaceli D, Rogaia D, Ardisia C, Romani R, Stangoni G, Pierini A, Donti E (2014) Recurrent approximately 100 Kb microdeletion in the chromosomal region 14q11.2, involving CHD8 gene, is associated with autism and macrocephaly. Am J Med Genet A 164A:3137–3141. https://doi.org/10.1002/ajmg.a.36741
Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, Walker MF, Ober GT, Teran NA, Song Y, El-Fishawy P, Murtha RC, Choi M, Overton JD, Bjornson RD, Carriero NJ, Meyer KA, Bilguvar K, Mane SM, Sestan N, Lifton RP, Gunel M, Roeder K, Geschwind DH, Devlin B, State MW (2012) De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485:237–241. https://doi.org/10.1038/nature10945
Stessman HA, Xiong B, Coe BP, Wang T, Hoekzema K, Fenckova M, Kvarnung M, Gerdts J, Trinh S, Cosemans N, Vives L, Lin J, Turner TN, Santen G, Ruivenkamp C, Kriek M, van Haeringen A, Aten E, Friend K, Liebelt J, Barnett C, Haan E, Shaw M, Gecz J, Anderlid BM, Nordgren A, Lindstrand A, Schwartz C, Kooy RF, Vandeweyer G, Helsmoortel C, Romano C, Alberti A, Vinci M, Avola E, Giusto S, Courchesne E, Pramparo T, Pierce K, Nalabolu S, Amaral DG, Scheffer IE, Delatycki MB, Lockhart PJ, Hormozdiari F, Harich B, Castells-Nobau A, Xia K, Peeters H, Nordenskjold M, Schenck A, Bernier RA, Eichler EE (2017) Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat Genet 49:515–526. https://doi.org/10.1038/ng.3792
Suetterlin P, Hurley S, Mohan C, Riegman KLH, Pagani M, Caruso A, Ellegood J, Galbusera A, Crespo-Enriquez I, Michetti C, Yee Y, Ellingford R, Brock O, Delogu A, Francis-West P, Lerch JP, Scattoni ML, Gozzi A, Fernandes C, Basson MA (2018) Altered neocortical gene expression, brain overgrowth and functional over-connectivity in Chd8 Haploinsufficient mice. Cereb Cortex 28:2192–2206. https://doi.org/10.1093/cercor/bhy058
Tavassoli T, Kolevzon A, Wang AT, Curchack-Lichtin J, Halpern D, Schwartz L, Soffes S, Bush L, Grodberg D, Cai G, Buxbaum JD (2014) De novo SCN2A splice site mutation in a boy with Autism spectrum disorder. BMC Med Genet 15:35. https://doi.org/10.1186/1471-2350-15-35
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164. https://doi.org/10.1093/nar/gkq603
Wang TY, Guo H, Xiong B, Stessman HAF, Wu HD, Coe BP, Turner TN, Liu YL, Zhao WJ, Hoekzema K, Vives L, Xia L, Tang MN, Ou JJ, Chen BY, Shen YD, Xun GL, Long M, Lin J, Kronenberg ZN, Peng Y, Bai T, Li HH, Ke XY, Hu ZM, Zhao JP, Zou XB, Xia K, Eichler EE (2016) De novo genic mutations among a Chinese autism spectrum disorder cohort. Nat Commun. https://doi.org/10.1038/Ncomms13316
Weiss K, Terhal PA, Cohen L, Bruccoleri M, Irving M, Martinez AF, Rosenfeld JA, Machol K, Yang Y, Liu P, Walkiewicz M, Beuten J, Gomez-Ospina N, Haude K, Fong CT, Enns GM, Bernstein JA, Fan J, Gotway G, Ghorbani M, van Gassen K, Monroe GR, van Haaften G, Basel-Vanagaite L, Yang XJ, Campeau PM, Muenke M (2016) De novo mutations in CHD4, an ATP-dependent chromatin remodeler gene, cause an intellectual disability syndrome with distinctive dysmorphisms. Am J Hum Genet 99:934–941. https://doi.org/10.1016/j.ajhg.2016.08.001
Whittaker DE, Riegman KL, Kasah S, Mohan C, Yu T, Sala BP, Hebaishi H, Caruso A, Marques AC, Michetti C, Smachetti ME, Shah A, Sabbioni M, Kulhanci O, Tee WW, Reinberg D, Scattoni ML, Volk H, McGonnell I, Wardle FC, Fernandes C, Basson MA (2017) The chromatin remodeling factor CHD7 controls cerebellar development by regulating reelin expression. J Clin Invest 127:874–887. https://doi.org/10.1172/JCI83408
Wu TJ, Shamsaddini A, Pan Y, Smith K, Crichton DJ, Simonyan V, Mazumder R (2014) A framework for organizing cancer-related variations from existing databases, publications and NGS data using a High-performance integrated virtual environment (HIVE). Database (Oxford). https://doi.org/10.1093/database/bau022
Acknowledgements
We appreciate the ASD participants and their families and also the physicians who took part in collection of samples. We also thank students (Yanhua Lv and Qing Ji) for their efforts on validation of CHD8 variants by Sanger sequencing in Yu An’s lab. We much appreciated Dr.Kai Wang from University of Pennsylvania who contributed to the annotation of frameshift variants of CHD8.
Funding
This work was supported by Grants from the Shanghai Municipal Science and Technology Major Project (Grant No. 2017SHZDZX01), National Natural Science Foundation of China (No. 31671386 and No. 91430112).
Author information
Authors and Affiliations
Contributions
YA, YSD, and YPS conceived the project and designed the experiments. LNZ, WWL and YSD collected ASD patients, performed autism behavior evaluation and clinical diagnosis as well as summarized the clinical information. YYJ and XC contributed one Autism patient. XPL also contributed one patient with mild intellectual disability with a CHD8 variant. QH and GL performed protein model analysis. JW and YA performed NGS analysis and variant calling. YA and her lab performed confirmation for the variants. YA and YPS did further analysis based on comprehensive database assessment (SSC and gnomAD). YA, JFG and YPS wrote and edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval
This study was approved by the Ethics Committee of Fudan University and informed consent was obtained from all families.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
An, Y., Zhang, L., Liu, W. et al. De novo variants in the Helicase-C domain of CHD8 are associated with severe phenotypes including autism, language disability and overgrowth. Hum Genet 139, 499–512 (2020). https://doi.org/10.1007/s00439-020-02115-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-020-02115-9