Abstract
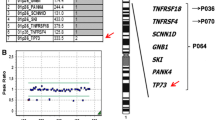
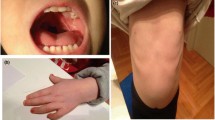
Terminal deletion in the short arm of chromosome 1 results in a disorder described as 1p36 deletion syndrome. The resulting phenotype varies among patients including mental retardation, developmental delay, sensorineural hearing loss, seizures, heart defects, and distinct facies. In the present case, we performed array-comparative genomic hybridization in a boy with multiple congenital malformations presenting some features overlapping the 1p36 deletion phenotype for whom chromosomal analysis did not reveal a terminal deletion in 1p. Results showed complex chromosome rearrangements involving the 1p36.33-p35.3 region. While the mechanism of origin of these rearrangements is still unclear, chromothripsis—a single catastrophic event leading to shattering chromosomes or chromosome regions and rejoining of the segments—has been described to occur in a fraction of cancers. The presence of at least 12 clustered breaks at 1p and apparent lack of mosaicism in the present case suggests that a single event like chromothripsis occurred. This finding suggests that chromothripsis is responsible for some constitutive complex chromosome rearrangements.


Similar content being viewed by others
References
Allemeersch J, Vooren VS, Hannes F, De Moor B, Vermeesch JR, Moreau Y (2009) An experimental loop design for the detection of constitutional chromosomal aberrations by array CGH. BMC Bioinform 10(1):380
Battaglia A (2011) Commentary: recognizing syndromes with overlapping features: how difficult is it? Considerations generated by the article on differential diagnosis of Smith–Magenis syndrome by Vieira and colleagues. Am J Med Genet Part A 155(5):986–987
Battaglia A, Hoyme HE, Dallapiccola B et al (2008) Further delineation of deletion 1p36 syndrome in 60 patients: a recognizable phenotype and common cause of developmental delay and mental retardation. Pediatrics 121:404–410
Chen E, Obolensky E, Rauen KA, Shaffer LG, Li X (2008) Cytogenetic and array CGH characterization of de novo 1p36 duplications and deletion in a patient with congenital cataracts, hearing loss, choanal atresia, and mental retardation. Am J Med Genet Part A 146A(21):2785–2790
Chen JM, Férec C, Cooper DN (2012) Transient hypermutability, chromothripsis and replication-based mechanisms in the generation of concurrent clustered mutations. Mutat Res 750:52e9
Gajecka M, Yu W, Ballif BC et al (2005) Delineation of mechanisms and regions of dosage imbalance in complex rearrangements of 1p36 leads to a putative gene for regulation of cranial suture closure. Eur J Hum Genet 13(2):139–149
Gajecka M, Mackay KL, Shaffer LG (2007) Monosomy 1p36 deletion syndrome. Am J Med Genet C Semin Med Genet 145C(4):346–356
Giannikou K, Fryssira H, Oikonomakis V, Syrmou A, Kosma K, Tzetis M, Kitsio-Tzeli S, Kanavakis E (2012) Further delineation of novel 1p36 rearrangements by array-CGH analysis: narrowing the breakpoints and clarifying the “extended” phenotype. Gene 506(2):360–368
Heilstedt HA, Ballif BC, Howard LA, Lewis RA, Stal S, Kashork CD, Bacino CA, Shapira SK, Shaffer LG (2003) Physical map of 1p36, placement of breakpoints in monosomy 1p36, and clinical characterization of the syndrome. Am J Hum Genet 72(5):1200–1212
Kloosterman WP, Guryev V, van Roosmalen M et al (2011) Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Hum Mol Genet 20(10):1916–1924
Liu P, Erez A, Nagamani SC et al (2011) Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell 146(6):889–903
Macera MJ, Sobrino A, Levy B et al (2015) Prenatal diagnosis of chromothripsis, with nine breaks characterized by karyotyping, FISH, microarray and whole-genome sequencing. Prenat Diagn 35(3):299–301
Pai GS, Thomas GH, Mahoney W, Migeon BR (1980) Complex chromosome rearrangements: report of a new case and literature review. Clin Genet 18(6):436–444
Pellestor F, Anahory T, Lefort G, Puechberty J, Liehr T, Hédon B, Sarda P (2011) Complex chromosomal rearrangements: origin and meiotic behavior. Hum Reprod Update 17(4):476–494
Plaisancié J, Kleinfinger P, Cances C, Bazin A, Julia S, Trost D, Lohmann L, Vigouroux A (2014) Constitutional chromoanasynthesis: description of a rare chromosomal event in a patient. Eur J Med Genet 57(10):567–570
Stephens PJ, Greenman CD, Fu B et al (2011) Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144(1):27–40
Vieira GH, Rodriguez JD, Boy R, de Paiva IS, DuPont BR, Moretti-Ferreira D, Srivastava AK (2011) Differential diagnosis of Smith–Magenis syndrome: 1p36 deletion syndrome. Am J Med Genet Part A 155:988–992
Zhang F, Carvalho CM, Lupski JR (2009) Complex human chromosomal and genomic rearrangements. Trends Genet 25(7):298–307
Acknowledgments
We thank the patient and his mother for their cooperation. Genetics analysis was supported by grants from the Fundacao de Amparo a Pesquisa do Estado de Sao Paulo (2010/18740-2 and 2011/07012-9) and Conselho Nacional de Desenvolvimento Cientifico e Tecnológico (306741/2012-1).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
Rights and permissions
About this article
Cite this article
Gamba, B.F., Richieri-Costa, A., Costa, S. et al. Chromothripsis with at least 12 breaks at 1p36.33-p35.3 in a boy with multiple congenital anomalies. Mol Genet Genomics 290, 2213–2216 (2015). https://doi.org/10.1007/s00438-015-1072-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-015-1072-0