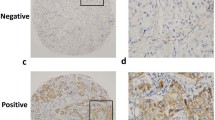
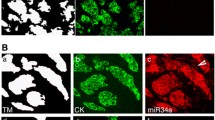
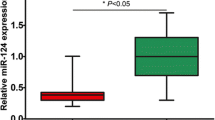
Abstract
Purpose
To visualize by in situ hybridization (ISH) the levels of a set of proliferation-associated miRNAs and to evaluate their impact and clinical applicability in prognostication of invasive breast carcinoma.
Methods
Tissue specimen from breast carcinoma patients were investigated for miRNAs-494, -205, -21 and -126. Prognostic associations for levels of miRNAs were analyzed based on complete clinical data and up to 22.5-year follow-up of the patient material (n = 285). For detection of the miRNAs, an automated sensitive protocol applying in situ hybridization was developed.
Results
MiRNA-494 indicated prognostic value for patients with invasive breast carcinoma. Among node-negative disease reduced level of miRNA-494 predicted 8.5-fold risk of breast cancer death (p = 0.04). Altered levels and expression patterns of the studied miRNAs were observed in breast carcinomas as compared to benign breast tissue.
Conclusions
The present paper reports for the first time on the prognostic value of miRNA-494 in invasive breast cancer. Particularly, detection of miRNA-494 could benefit patients with node-negative breast cancer in identifying subgroups with aggressive disease. Based on our experience, the developed automatic ISH method to visualize altered levels of miRNAs-494, -205, -21 and -126 could be applied to routine pathology diagnostics providing that conditions of tissue treatment, especially fixation delays, are managed.



Similar content being viewed by others
References
Baranwal S, Alahari SK (2010) miRNA control of tumor cell invasion and metastasis. Int J Cancer 126:1283–1290
Carleton M, Cleary MA, Linsley PS (2007) MicroRNAs and cell cycle regulation. Cell Cycle 6:2127–2132
Chatterjee N, Rana S, Espinosa-Diez C, Anand S (2017) MicroRNAs in cancer: challenges and opportunities in early detection, disease monitoring, and therapeutic agents. Curr Pathobiol Rep 5:35–42
Childs G, Fazzari M, Kung G et al (2009) Low-level expression of microRNAs let-7d and miR-205 are prognostic markers of head and neck squamous cell carcinoma. Am J Pathol 174:736–745
Chivukula RR, Mendell JT (2008) Circular reasoning: microRNAs and cell-cycle control. Trends Biochem Sci 33:474–481
Chugh P, Dittmer DP (2012) Potential pitfalls in microRNA profiling. Wiley Interdiscip Rev RNA 3:601–616
Duffy MJ, Walsh S, McDermott EW, Crown J (2015) Biomarkers in breast cancer: where are we and where are we going? Adv Clin Chem 71:1–23
Ebrahimi F, Gopalan V, Smith RA, Lam AK (2014) miR-126 in human cancers: clinical roles and current perspectives. Exp Mol Pathol 96:98–107
Goldhirsch A, Ingle JN, Gelber RD et al (2009) Thresholds for therapies: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer. Ann Oncol 20:1319–1329
Goldhirsch A, Wood WC, Coates AS et al (2011) Strategies for subtypes—dealing with the diversity of breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer. Ann Oncol 22:1736–1747
Greene SB, Herschkowitz JI, Rosen JM (2010) The ups and downs of miR-205: identifying the roles of miR-205 in mammary gland development and breast cancer. RNA Biol 7:300–304
Gurvits N, Repo H, Löyttyniemi E, Nykänen M, Anttinen J, Kuopio T, Talvinen K, Kronqvist P (2016) Prognostic implications of securin expression and sub-cellular localization in human breast cancer. Cell Oncol (Dordr) 39:319–331
Haga CL, Phinney DG (2012) MicroRNAs in the imprinted DLK1-DIO3 region repress the epithelial-to-mesenchymal transition by targeting the TWIST1 protein signaling network. J Biol Chem 287:42695–42707
Hagman Z, Haflidadóttir BS, Ceder JA et al (2013) miR-205 negatively regulates the androgen receptor and is associated with adverse outcome of prostate cancer patients. Br J Cancer 108:1668–1676
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144:646–674
Hanna JA, Hahn L, Agarwal S et al (2012) In situ measurement of miR-205 in malignant melanoma tissue supports its role as a tumor suppressor microRNA. Lab Investig 92:1390–1397
He W, Li Y, Chen X, Lu L, Tang B, Wang Z, Pan Y, Cai S, He Y, Ke Z (2014) miR-494 acts as an anti-oncogene in gastric carcinoma by targeting c-myc. J Gastroenterol Hepatol 29:1427–1434
Healey MA, Hirko KA, Beck AH, Collins LC, Schnitt SJ, Eliassen AH, Holmes MD, Tamimi RM, Hazra A (2017) Assessment of Ki67 expression for breast cancer subtype classification and prognosis in the Nurses’ Health Study. Breast Cancer Res Treat 166:613–622
Hou SX, Ding BJ, Li HZ et al (2013) Identification of microRNA-205 as a potential prognostic indicator for human glioma. J Clin Neurosci 20:933–937
Hug KA, Anthony L, Eldeiry D, Benson J, Wheeler E, Mousa S, Shi B (2015) Expression and tissue distribution of MicroRNA-21 in malignant and benign breast tissues. Anticancer Res 35:3175–3183
Hulf T, Sibbritt T, Wiklund ED et al (2013) Epigenetic-induced repression of microRNA-205 is associated with MED1 activation and a poorer prognosis in localized prostate cancer. Oncogene 32:2891–2899
Huo L, Wang Y, Gong Y, Krishnamurthy S, Wang J, Diao L, Liu CG, Liu X, Lin F, Symmans WF, Wei W, Zhang X, Sun L, Alvarez RH, Ueno NT, Fouad TM, Harano K, Debeb BG, Wu Y, Reuben J, Cristofanilli M, Zuo Z (2016) MicroRNA expression profiling identifies decreased expression of miR-205 in inflammatory breast cancer. ModPathol 29:330–346
Jena MK (2017) MicroRNAs in the development and neoplasia of the mammary gland. F1000Res 6:1018
Jørgensen S, Baker A, Møller S, Nielsen BS (2010) Robust one-day in situ hybridization protocol for detection of microRNAs in paraffin samples using LNA probes. Methods 52:375–381
Kaboli PJ, Rahmat A, Ismail P, Ling KH (2015) MicroRNA-based therapy and breast cancer: a comprehensive review of novel therapeutic strategies from diagnosis to treatment. Pharmacol Res 97:104–121
Kalogirou C, Spahn M, Krebs M, Joniau S, Lerut E, Burger M, Scholz CJ, Kneitz S, Riedmiller H, Kneitz B (2013) MiR-205 is progressively down-regulated in lymph node metastasis but fails as a prognostic biomarker in high-risk prostate cancer. Int J Mol Sci 14:21414–21434
Kapanidou M, Curtis NL, Bolanos-Garcia VM (2017) Cdc20: at the crossroads between chromosome segregation and mitotic exit. Trends Biochem Sci 42:193–205
Karaayvaz M, Zhang C, Liang S et al (2013) Prognostic significance of miR-205 in endometrial cancer. PLoS One 7:e35158
Karra H, Pitkänen R, Nykänen M, Talvinen K, Kuopio T, Söderström M, Kronqvist P (2012) Securin predicts aneuploidy and survival in breast cancer. Histopathology 60:586–596
Karra H, Repo H, Ahonen I, Löyttyniemi E, Pitkänen R, Lintunen M, Kuopio T, Söderström M, Kronqvist P (2014) Cdc20 and securin overexpression predict short-term breast cancer survival. Br J Cancer 110:2905–2913
Kim WK, Park M, Kim YK, Tae YK, Yang HK, Lee JM, Kim H (2011) MicroRNA-494 downregulates KIT and inhibits gastrointestinal stromal tumor cell proliferation. Clin Cancer Res 17:7584–7594
Lim L, Balakrishnan A, Huskey N, Jones KD, Jodari M, Ng R, Song G, Riordan J, Anderton B, Cheung ST, Willenbring H, Dupuy A, Chen X, Brown D, Chang AN, Goga A (2014) MicroRNA-494 within an oncogenic microRNA megacluster regulates G1/S transition in liver tumorigenesis through suppression of mutated in colorectal cancer. Hepatology 59:202–215
Liu K, Liu S, Zhang W, Jia B, Tan L, Jin Z, Liu Y (2015) miR-494 promotes cell proliferation, migration and invasion, and increased sorafenib resistance in hepatocellular carcinoma by targeting PTEN. OncolRep 34:1003–1010
Ma YB, Li GX, Hu JX, Liu X, Shi BM (2015) Correlation of miR-494 expression with tumor progression and patient survival in pancreatic cancer. Genet Mol Res 14:18153–18159
MacKenzie TA, Schwartz GN, Calderone HM, Graveel CR, Winn ME, Hostetter G, Wells WA, Sempere LF (2014) Stromal expression of miR-21 identifies high-risk group in triple-negative breast cancer. Am J Pathol 184:3217–3225
Marino AL, Evangelista AF, Vieira RA, Macedo T, Kerr LM, Abrahão-Machado LF, Longatto-Filho A, Silveira HC, Marques MM (2014) MicroRNA expression as risk biomarker of breast cancer metastasis: a pilot retrospective case-cohort study. BMC Cancer 14:739
Markou A, Yousef GM, Stathopoulos E et al (2014) Prognostic significance of metastasis-related microRNAs in early breast cancer patients with a long follow-up. Clin Chem 60:197–205
Nicolini A, Ferrari P, Duffy MJ (2017) Prognostic and predictive biomarkers in breast cancer: past, present and future. Semin Cancer Biol. https://doi.org/10.1016/j.semcancer.2017.08.010
Nielsen BS, Balslev E, Poulsen TS, Nielsen D, Møller T, Mortensen CE, Holmstrøm K, Høgdall E (2014) miR-21 expression in cancer cells may not predict resistance to adjuvant trastuzumab in primary breast cancer. Front Oncol 4:207
Piovan C, Palmieri D, Di Leva G, Braccioli L, Casalini P, Nuovo G, Tortoreto M, Sasso M, Plantamura I, Triulzi T, Taccioli C, Tagliabue E, Iorio MV, Croce CM (2012) Oncosuppressive role of p53-induced miR-205 in triple negative breast cancer. Mol Oncol 6:458–472
Pritchard CC, Cheng HH, Tewari M (2012) MicroRNA profiling: approaches and considerations. Nat Rev Genet 13:358–369
Qi L, Bart J, Tan LP, Platteel I, Sluis T, Huitema S, Harms G, Fu L, Hollema H, Berg A (2009) Expression of miR-21 and its targets (PTEN, PDCD4, TM1) in flat epithelial atypia of the breast in relation to ductal carcinoma in situ and invasive carcinoma. BMC Cancer 9:163-2407-9-163
Quesne JL, Jones J, Warren J, Dawson SJ, Ali HR, Bardwell H, Blows F, Pharoah P, Caldas C (2012) Biological and prognostic associations of miR-205 and let-7b in breast cancer revealed by in situ hybridization analysis of micro-RNA expression in arrays of archival tumour. J Pathol 227:306–314
R Core Team (2017) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Ranganathan K, Sivasankar V (2014) MicroRNAs—biology and clinical applications. J Oral Maxillofac Pathol 18:229–234
Rask L, Balslev E, Jorgensen S, Eriksen J, Flyger H, Moller S, Hogdall E, Litman T, Nielsen BS (2011) High expression of miR-21 in tumor stroma correlates with increased cancer cell proliferation in human breast cancer. APMIS 119:663–673
Repo H, Gurvits N, Löyttyniemi E, Nykänen M, Lintunen M, Karra H, Kurki S, Kuopio T, Talvinen K, Söderström M, Kronqvist P (2017) PTTG1-interacting protein (PTTG1IP/PBF) predicts breast cancer survival. BMC Cancer 17:705
Sempere LF (2014) Fully automated fluorescence-based four-color multiplex assay for co-detection of microRNA and protein biomarkers in clinical tissue specimens. Methods Mol Biol 1211:151–170
Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, Wells W, Kauppinen S, Cole CN (2007) Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res 67:11612–11620
Sempere LF, Preis M, Yezefski T, Ouyang H, Suriawinata AA, Silahtaroglu A, Conejo-Garcia JR, Kauppinen S, Wells W, Korc M (2010) Fluorescence-based codetection with protein markers reveals distinct cellular compartments for altered MicroRNA expression in solid tumors. Clin Cancer Res 16:4246–4255
Singh U, Keirstead N, Wolujczyk A, Odin M, Albassam M, Garrido R (2014) General principles and methods for routine automated microRNA in situ hybridization and double labeling with immunohistochemistry. Biotech Histochem 89:259–266
Song L, Liu D, Wang B, He J, Zhang S, Dai Z, Ma X, Wang X (2015) miR-494 suppresses the progression of breast cancer in vitro by targeting CXCR4 through the Wnt/β-catenin signaling pathway. Oncol Rep 34:525–531
Sun HB, Chen X, Ji H, Wu T, Lu HW, Zhang Y, Li H, Li YM (2014) miR 494 is an independent prognostic factor and promotes cell migration and invasion in colorectal cancer by directly targeting PTEN. Int J Oncol 45:2486–2494
Talvinen K, Karra H, Hurme S, Nykänen M, Nieminen A, Anttinen J, Kuopio T, Kronqvist P (2009) Securin promotes the identification of favourable outcome in invasive breast cancer. Br J Cancer 101:1005–1010
Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J (2008) Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451:147–152
Urbanek MO, Nawrocka AU, Krzyzosiak WJ (2015) Small RNA detection by in situ hybridization methods. Int J Mol Sci 16(6):13259–13286
Yamanaka S, Campbell NR, An F, Kuo SC, Potter JJ, Mezey E, Maitra A, Selaru FM (2012) Coordinated effects of microRNA-494 induce G(2)/M arrest in human cholangiocarcinoma. CellCycle 11:2729–2738
Yang YK, Xi WY, Xi RX, Li JY, Li Q, Gao YE (2015) MicroRNA-494 promotes cervical cancer proliferation through the regulation of PTEN. OncolRep 33:2393–2401
Yang A, Wang X, Yu C, Jin Z, Wei L, Cao J, Wang Q, Zhang M, Zhang L, Zhang L, Hao C (2017) microRNA-494 is a potential prognostic marker and inhibits cellular proliferation, migration and invasion by targeting SIRT1 in epithelial ovarian cancer. Oncol Lett 3177–3184
Yerushalmi R, Woods R, Ravdin PM, Hayes MM, Gelmon KA (2010) Ki67 in breast cancer: prognostic and predictive potential. Lancet Oncol 11:174–183
Zhang J, Du YY, Lin YF, Chen YT, Yang L, Wang HJ, Ma D (2008) The cell growth suppressor, mir-126, targets IRS-1. Biochem Biophys Res Commun 377:136–140
Zhang JY, Sun MY, Song NH, Deng ZL, Xue CY, Yang J (2015) Prognostic role of microRNA-205 in multiple human malignant neoplasms: a meta-analysis of 17 studies. BMJ Open 5(1):e006244
Zhang C, Liu K, Li T, Fang J, Ding Y, Sun L, Tu T, Jiang X, Du S, Hu J, Zhu W, Chen H, Sun X (2016) miR-21: a gene of dual regulation in breast cancer. Int J Oncol 48:161–172
Zhao X, Zhou Y, Chen YU, Yu F (2016) miR-494 inhibits ovarian cancer cell proliferation and promotes apoptosis by targeting FGFR2. OncolLett 11:4245–4251
Acknowledgements
The authors thank Mrs. Sinikka Collanus, PhD Markus Peurla and Mr. Jaakko Liippo, Institute of Biomedicine, University of Turku, Turku, Finland.
Funding
The study was supported by grants from Cancer Society of South-West Finland, Foundation of Finska Läkaresällskapet and Turku University Hospital.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The work has been approved by the ethics committees of Turku University Hospital and Central Hospital of Central Finland, Turku, Finland (permit numbers 0286/2002, 7765-2002 and AB159859). All patient material has been obtained from archives of biobanks and includes written consent from patients obtained in accordance to the ethics permits and the Finnish Biobank Act (688/2012).
Rights and permissions
About this article
Cite this article
Gurvits, N., Autere, TA., Repo, H. et al. Proliferation-associated miRNAs-494, -205, -21 and -126 detected by in situ hybridization: expression and prognostic potential in breast carcinoma patients. J Cancer Res Clin Oncol 144, 657–666 (2018). https://doi.org/10.1007/s00432-018-2586-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-018-2586-8