Abstract
Pectin is one of the main components of the plant cell wall that functions as the primary barrier against pathogens. Among the extracellular pectinolytic enzymes, pectin methylesterase (PME) demethylesterifies pectin, which is secreted into the cell wall in a highly methylesterified form. Here, we isolated and functionally characterized the pepper (Capsicum annuum L.) gene CaPMEI1, which encodes a pectin methylesterase inhibitor protein (PMEI), in pepper leaves infected by Xanthomonas campestris pv. vesicatoria (Xcv). CaPMEI1 transcripts are localized in the xylem of vascular bundles in leaf tissues, and pathogens and abiotic stresses can induce differential expression of this gene. Purified recombinant CaPMEI1 protein not only inhibits PME, but also exhibits antifungal activity against some plant pathogenic fungi. Virus-induced gene silencing of CaPMEI1 in pepper confers enhanced susceptibility to Xcv, accompanied by suppressed expression of some defense-related genes. Transgenic Arabidopsis CaPMEI1-overexpression lines exhibit enhanced resistance to Pseudomonas syringae pv. tomato, mannitol and methyl viologen, but not to the biotrophic pathogen Hyaloperonospora parasitica. Together, these results suggest that CaPMEI1, an antifungal protein, may be involved in basal disease resistance, as well as in drought and oxidative stress tolerance in plants.
Similar content being viewed by others
Introduction
Plant cell wall, the first barrier of defense against invading pathogens, is composed of cellulose microfibrils cross-linked by hemicellulose, pectin and extensin. Pectin comprises a highly heterogeneous group of polymers that includes homogalacturonans and rhamnogalacturonans I and II (Willats et al. 2001a, b). In pectin polymers, the galacturonic acid carboxyl groups can be methylesterified by a group of pectinases. These galacturonic acid methylesters are hydrolyzed by pectin methylesterase (PME) (Hagerman and Austin 1986; Pelloux et al. 2007). Many physiological processes, such as fruit maturation, microsporangenesis, seed germination and pollen growth, are affected by the degree to which PME changes the methlyesterification of galacturonic acid (Tieman and Handa 1994; Kagan-Zur et al. 1995; Ren and Kermode 2000; Bosch et al. 2005). The number and distribution of free and unesterified galacturonate carboxyl groups along the homogalacturonan chain has a great influence on the pectin properties and cell wall firmness (Willats et al. 2001a, b).
Plant pathogens hydrolyze the cell wall components of plants using extracellular pectinolytic enzymes (Collmer and Keen 1986), and PME is found in many plant pathogenic bacteria and fungi (Asoufi et al. 2007). The black filamentous fungus Aspergillus niger secretes a set of pectin-degrading enzymes that include PME, polygalacturonase and pectin lyase, and these decompose the plant cell wall to establish infection and absorb nutrients from the host (de Vries and Visser 2001). In soft rot disease caused by Erwinia chrysanthemi, bacterial PME activity is induced during infection of Saintpaulia plants; however, PME-deficient mutants are noninvasive to the host cells (Boccara and Chatain 1989). PMEs have also been found in higher plants; they play significant roles in physiological processes and interactions with pathogens (Micheli 2001). In tobacco plants, host cell pectin methylesterases are required for the tobacco mosaic virus movement protein to transfer the viruses between host cells (Chen et al. 2000). Furthermore, PME-degraded polygalacturonans are associated with recognition of fungal pathogens (Wietholter et al. 2003). PME is also involved in symbiosis-specific functions (Lievens et al. 2002). For example, plant PME isoenzymes may undergo organism-specific post-translational processing for structural and functional integrity during interactions with various microorganisms (Micheli 2001).
The PME enzyme activity is modulated specifically by inhibitor proteins such as the pectin methylesterase inhibitor (PMEI; Micheli 2001). Moreover, the PMEIs that inhibit demethylesterification of highly heterogeneous polymers (pectins) are the plant invertase inhibitor-related proteins, which are inhibitors of important metabolic enzymes (Koch 1996). Plant invertase inhibitor-related proteins play key roles in wounding, the plant defense reaction and developmental transitions (Raush and Greiner 2004), as well as during osmotic stress, senescence and seed development (Greiner et al. 1998, 1999). Investigation of gain- and loss-of-function mutants of tobacco cell wall invertase inhibitor (NtCIF) protein demonstrated that these inhibitor proteins play a role in seed development (Raush et al. 1998). Overexpression of the tobacco vacuolar invertase inhibitor protein (NtVIF) gene in transgenic potatoes is of potential use in the field of food technology (Greiner et al. 1998). However, there is little known about the in vivo functions of PMEI protein.
Proteinaceous inhibitors have been purified from kiwi (Actinidia deliciosa) (Giovane et al. 2004), Arabidopsis (Wolf et al. 2003; Raiola et al. 2004), rice (Han et al. 2005) and the jelly fig (Ficus awkeotsang cv. Makino) (Jiang et al. 2001, 2002). The kiwi PMEI is specific for PME (Balestrieri et al. 1990) and is active against PMEs from several plants, including kiwi, orange, apple, tomato, apricot, carrot, potato and banana (Ly-Nguyen et al. 2004). Four Cys residues conserved in several isoforms of PMEI are involved in the formation of disulfide bridges (Camardella et al. 2000). PME and PMEI form a stoichiometric 1:1 complex, in which the interaction between the PME and the inhibitor occurs in close proximity to the putative active site (Di Matteo et al. 2005). Since PME activity can be modulated by pH, the stability of the PME–PMEI complex is also affected by pH (Denès et al. 2000). Crystallographic work has revealed that an α-helical hairpin motif plays a structurally important role in PMEI activation (Hothorn et al. 2004). Many cDNAs encoding PMEIs have been isolated and functionally characterized from plants (Rausch and Greiner 2004); however, their role in plant defense remains relatively unknown.
To date, the functional analyses of genes associated with defense responses in plants have utilized reverse-genetics approaches based on loss-of-function via double-stranded RNA interference (Robertson 2004) or gain-of-function via transgenic gene expression (Clough and Bent 1998). Virus-induced gene silencing (VIGS) has been proven to be a useful method for assessing the function of target genes in Solanum species (Brigneti et al. 2004). In particular, VIGS studies have been used to investigate disease resistance signaling and defense-related genes such as SGT1 (Liu et al. 2002c; Peart et al. 2002b), EDS1 (Liu et al. 2002b; Peart et al. 2002a) and NPR1/NIM1 (Liu et al. 2002b) in Nicotiana benthamiana. AtPGIP1 is among the genes encoding pectic enzyme-related proteins; it encodes polygalacturonase (PG)-inhibiting protein (PGIP), and an antisense AtPGIP1 gene was used to silence its expression in transgenic Arabidopsis plants (Ferrari et al. 2006). Previous studies have revealed that overexpression of two closely related genes, AtPGIP1 and AtPGIP2, conferred resistance against Botrytis cinerea infection (Ferrari et al. 2003). Silencing of AtPGIP1 resulted in enhanced susceptibility to infection, as well as reduced activity of PGIP (Ferrari 2006). However, gene-silencing techniques such as VIGS and antisense RNA have not yet been used to investigate the role played in plant defense by other PMEI-encoding genes.
Here, we used a macroarray technique to isolate and functionally characterize a pectin methylesterase inhibitor gene, CaPMEI1, from a cDNA library of pepper (Capsicum annuum L.) leaves infected with Xanthomonas campestris pv. vesicatoria (Xcv; Jung and Hwang 2000). Local and systemic induction of CaPMEI1 was investigated in pepper plants following inoculation with pathogenic and non-pathogenic bacteria. We also examined the involvement of CaPMEI1 in defense-related signal transduction cascades via exogenous application of abiotic elicitors to pepper plants. Recombinant CaPMEI1 proteins were expressed in E. coli and exhibited antifungal activity against plant pathogenic fungi. Since it is difficult to transform pepper plants, we performed gene silencing and CaPMEI1 overexpression in pepper and Arabidopsis, respectively, to identify the cellular functions of the CaPMEI1 gene. The functional data obtained by VIGS and transgenic ectopic expression of CaPMEI1 suggest that this pepper pectin methylesterase inhibitor is involved in plant defense and abiotic stress responses.
Materials and methods
Plant materials and growth conditions
Pepper (Capsicum annuum L. cv. Nockwang) plants were grown at 28°C under a 16 h day at 70 μmol photons m−2 s−1. Plants were seeded into a plastic tray (55 × 35 × 15 cm3) containing steam-sterilized soil mix (peat moss, perlite and vermiculite; 5:3:2, v/v/v) and loam soil (1:1, v/v). At the two-leaf stage, seedlings were transplanted into plastic pots (5 × 15 × 10 cm3) containing the soil mix previously described.
Arabidopsisthaliana (ecotype Columbia) plants were grown in pots containing vermiculite, peat moss and perlite (1:1:0.5, v/v/v) in a growth chamber under a 12 h light/12 h dark photoperiod (130 μmol photons m−2 s−1) at 24°C and 60% relative humidity. Prior to sowing, the seeds were surface-sterilized using 1% sodium hypochlorite and vernalized at 4°C for 3 days to break dormancy.
Pathogens, inoculation procedures, disease rating and tissue staining
Xanthomonas campestris pv. vesicatoria (Xcv) strains Ds1 and Bv5-4a were used in this study. Bacteria were cultured overnight in yeast-nutrient (YN) broth (5 g L−1 yeast extract, 8 g L−1 nutrient broth) at 28°C. Prior to inoculation, bacteria were harvested by centrifugation and resuspended in sterile tap water (108 cfu mL−1). Pepper plants were inoculated at the six-leaf stage by infiltrating the bacterial suspension into the abaxial side of fully expanded leaves using an atomizer. Infected plants were then incubated for 16 h at 28°C in a moist chamber with 100% relative humidity. Bacteria-infected leaves were sampled at various time intervals after inoculation. To evaluate systemic induction in the upper leaves, bacterial suspensions (108 cfu ml−1) were infiltrated into the lower leaves of pepper plants at the two-leaf stage using a needless syringe. The bacterial strains used for this study included: virulent and avirulent strains of X. campestris pv. vesicatoria (Ds1 and Bv5-4a); a non-pathogenic strain (Pseudomonasfluorescens ATCC13525); and Escherichiacoli JM109.
Leaves of 6-week-old Arabidopsis plants were infiltrated with a suspension (OD600 = 0.001) of virulent Pseudomonas syringae pv. tomato strain DC3000 (Pst DC3000). The bacteria were cultured overnight at 28°C and suspended in 10 mM MgCl2. To determine bacterial growth, leaf discs were cut from infected leaves at different time intervals after inoculation. Bacterial growth was monitored by performing serial dilutions onto KB agar containing 100 μg mL−1 rifampicin. Each experiment was replicated three times.
Hyaloperonospora parasitica isolate Noco2 was propagated by weekly subculturing on 7- to 10-day-old Arabidopsis seedlings. The 7-day-old seedlings were inoculated with an H. parasitica asexual inoculum (5 × 104 conidiosporangia mL−1). The seedlings inoculated with H. parasitica were covered with a plastic dome to maintain a high relative humidity (80–100%) and grown in a growth chamber at 17°C. Seven days after inoculation, disease rating was scored for more than 50 plants per treatment. A visual disease rating was expressed as the number of sporangiophores on each cotyledon and was divided into five classes: 0–5, 6–10, 11–15, 16–20 and >20 sporangiophores per cotyledon. The cotyledons from inoculated plants were stained with lactophenol-trypan blue (10 mL lactic acid, 10 mL glycerol, 10 g phenol and 10 mg trypan blue, dissolved in 10 mL of distilled water) to assess H. parasitica growth. At 2–5 days after inoculation, the infected cotyledons were boiled for 5 min in the staining solution and de-stained overnight in chloral hydrate (2.5 g chloral hydrate dissolved in 1 mL distilled water). The destained cotyledons were subsequently mounted in 70% glycerol for microscopic observation.
Isolation and sequence analysis of pathogen-induced cDNAs
To construct a pathogen-induced cDNA library, pepper leaves were inoculated with the avirulent strain X. campestris pv. vesicatoria Bv5-4a. The pathogen-induced cDNA library was constructed using 5 μg poly(A)+ mRNA extracted from inoculated pepper leaves (Kim and Hwang 2000). To isolate pathogen-inducible cDNAs from the pepper cDNA library, we performed differential hybridization, as described previously by Jung and Hwang (2000). Digoxigenin (DIG)-labeled, single-stranded cDNA probes were generated from total RNA of healthy and Bv5-4a-infected leaves using RT-PCR. Nylon membranes were pre-hybridized at 65°C for 3 h in 5× SSC, 0.1% sodium lauroylsarcosine, 0.02% SDS and 1% blocking reagent (Boehringer Mannheim, Mannheim, Germany). Hybridization was then performed overnight at 65°C in the same buffer with single-stranded cDNA probes. Hybridized membranes were rinsed twice for 5 min with 2× SSC and 0.1% SDS at room temperature, and twice for 10 min with 0.1× SSC and 0.1% SDS at 65°C. The hybridization signals were detected according to the manufacturer’s protocol (Boehringer Mannheim).
We selected cDNA clones that were expressed strongly in pathogen-infected leaves, compared with those of healthy leaves. Clones were sequenced with an ABI 310 DNA sequencer (Applied Biosystems, Foster City, CA, USA) using the PRISM Big Dye™ Terminator Cycle Sequencing Ready Reaction Kit (PE Biosystems, Foster City, CA, USA). Sequencing results were analyzed using BLAST (National Center for Biotechnology Information; Altschul et al. 1997).
Treatment with abiotic elicitors and environmental stresses
The leaves of pepper plants at the six-leaf stage were sprayed with 5 mM salicylic acid (SA), 100 μM methyl jasmonate (MeJA) or 100 μM absicisic acid (ABA). Pepper plants treated with methyl jasmonate were sealed tightly in plastic bags. For ethylene treatment, whole plants were removed from soil, and then placed in a water-containing glass chamber, followed by injection of ethylene gas (5 μL L−1). For cold stress treatment, plants were placed at 4°C in a cold room. For wounding stress, the leaves were pricked with needles. To impose drought stress, the plants were removed from the soil and then incubated at room temperature without water. H2O2 treatment was performed by spraying leaves with 100 mM H2O2 solution. Leaves treated with various elicitors and abiotic stresses were removed from the plants, frozen in liquid nitrogen and stored at −70°C until used for RNA isolation.
RNA isolation and RNA gel blot analysis
Total RNA was extracted from pepper leaves, stems, roots, flowers and fruits using the guanidine isothiocyanate method (Chomczynski and Sacchi 1987). Frozen tissues (1 g) were ground to a powder and homogenized in 10 mL extraction buffer (4 M guanidine isothiocyanate, 25 mM sodium citrate [pH 7.0], 0.55% [w/v] N-laurylsarcosine and 0.1 M 2-mercaptoethanol). A mixture of 2 M sodium acetate (pH 4.0), water-saturated phenol and chloroform–isoamylalcohol (24:1) was added to the homogenate, followed by precipitation. Total RNA (20 μg) was separated by 1.2% formaldehyde-agarose gel electrophoresis and then blotted onto Hybond-N+ membranes (Amersham, Buckinghamshire, UK). Transferred RNA was fixed to the membrane using UV cross-linking.
The 3′ UTR region of CaPMEI1 was amplified for use in the generation of a gene-specific probe. The primers used for amplification of the CaPMEI1 gene-specific region were 5′-CATGGGTAAGTGCTGCCTTGACGGAC-3′ and 5′-GTTAACAAATGCATA TGGAACATTT-3′. The CaBPR1 coding region was amplified with the primers 5′-ATGGGACACTCTAATATTGCC-3′ and 5′-GACATCAGTTGGAAGTTCCAA-3′. The CaSAR82A coding region was amplified using the primers 5′-ATGGTTTCCAAAAGT AGTATTTTTATTT-3′ and 5′-TATGCTTAACAATTATTACTGAATA TAATC-3′. PCR-amplified products were 32P-labeled using a random priming kit (Boehringer, Mannheim). Hybridization was performed overnight at 65°C in 5% dextran sulfate, 0.25 M disodium phosphate (pH 7.2), 7% (w/v) sodium dodecyl sulfate (SDS) and 1 mM EDTA. Following hybridization, the membranes were washed twice for 10 min with 2× SSC and 0.1% SDS at room temperature, and then twice for 5 min with 0.1× SSC and 0.1% SDS at 65°C. Equal loading of RNA was confirmed by ethidium bromide-staining of ribosomal RNA.
In situ RNA localization
In situ RNA localization was performed as described previously (Lee et al. 2000). Leaf tissue was fixed for 2 h in a solution of 1× phosphate-buffered saline (PBS), 4% paraformaldehyde and 1 μL mL−1 Triton X-100 by vacuum infiltration for 10 min and shaking for 2 h at room temperature. The fixed samples were washed with 1× PBS, dehydrated through a graded ethanol and xylol series, and then embedded in liquid paraplast at 57°C (Sherwood Medical, St. Louis, MO, USA). Paraplast-embedded sections (10 μm in thickness) were placed on glass slides coated with poly-l-lysine (Sigma, St. Louis, MO, USA) and incubated at 42°C. Sections were deparafinated using xylene and 1-propanol, followed by rehydration with serial dilutions of ethanol. Tissue samples were treated with 0.01 M Tris–HCl (pH 8.0) and 1% bovine serum albumin (BSA) for 10 min, followed by incubation in 100 mM Tris–HCl solution (pH 8.0) containing proteinase K (5 mg mL−1) and 50 mM EDTA, for 30 min at 37°C. Sections were treated with 0.25% acetic anhydride in 100 mM triethanolamine (pH 8.0) for 10 min at room temperature to inhibit non-specific signals. Digoxigenin (DIG)-labeled probes were prepared using the Dig High Prime DNA Labeling and Detection kit, according to the manufacturer’s instructions (Boehringer Mannheim). Sections were prehybridized and hybridized at 42°C in 50% formamide, 4× SSC, 150 μg mL−1 tRNA and 0.5% blocking reagent (Boehringer Mannheim). After hybridization, the sections were washed twice with 50% formamide and 4× SSC at 42°C, twice with 4× SSC and then once with diethyl pyrocarbonate (DEPC)-treated water. The DIG signal was detected, according to the manufacturer’s instructions (Boehringer Mannheim). Color reactions were developed overnight at 37°C with nitro-blue tetrazolium chloride (NBT) and 5-bromo-4-chloro-3-indolyl phosphate (BCIP), and reactions were stopped with TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH 8.0). Sections were photographed with Kodak ISO 100 film under a bright field Olympus BH-2 microscope (Olympus, Tokyo, Japan). To demonstrate the specificity of in situ hybridization, control hybridizations were performed without DIG-labeled probes.
Purification of recombinant CaPMEI1 protein
The CaPMEI1 coding region, including the stop codon, was amplified by PCR using the forward and reverse primers 5′-GAATTCATGGAAGGTGGCAATTTTCT-3′ and 5′-CTCGAGTAGCCGTGAAGGGCAGCCAGACG-3′, respectively. Amplification products were cloned into pCR2.1-TOPO (Invitrogen, Carlsbad, CA, USA), which was digested with EcoRI and XhoI and ligated into the similarly digested pET32a (Novagen, Madison, WI, USA).
Escherichia coli BL21 (DE3) pLysS (Novagen), which is defective for thioredoxin reductase, was used as a host for recombinant protein expression. Cultures were started from single colonies, grown in LB broth at 37°C, and then diluted 1:100 at OD600 = 0.6. After dilution, bacteria were grown to a density of OD600 = 0.6, induced with 10 mM IPTG and grown for a further 5 h at 37°C. Cells were harvested by centrifugation for 15 min at 5,000g, extracted with denaturation buffer (8 M urea, 20 mM sodium phosphate buffer [pH 7.8], 500 mM NaCl) and disrupted by sonication. Following centrifugation at 5,000g for 15 min at 4°C, the supernatant was loaded onto a 1.5 mL column of Ni-NTA resin (Qiagen, Hilden, Germany), which was washed with an initial denaturing wash buffer (8 M urea, 20 mM sodium phosphate buffer [pH 6.0], 500 mM NaCl), followed by a second denaturing wash buffer (8 M urea, 20 mM sodium phosphate buffer [pH 5.3], 500 mM NaCl). Bound fusion protein was eluted with a final denaturing elution buffer (8 M urea, 20 mM sodium phosphate buffer [pH 4.0], 500 mM NaCl). Recombinant proteins were dialyzed against a buffer containing 10 mM Tris (pH 8.0) and 0.1% Triton X-100, according to the manufacturer’s protocol. As a control, thioredoxin was purified using a native buffer that did not contain urea. The purified CaPMEI1 recombinant protein was digested for 16 h at room temperature with recombinant enterokinase (1 U/5 μg recombinant protein; Novagen).
SDS polyacrylamide gel electrophoresis
CaPMEI1 was dissolved in 1× SDS sample buffer (0.9 g glycerol, 5% SDS, 1% bromophenol blue, 0.1 mL mercaptoethanol and 1 L H2O), separated by 12% SDS-PAGE as described previously (Laemmli 1970), and stained with Coomassie Brilliant Blue R-250. Molecular weights were estimated using 6.5 to 200.5 kDa marker proteins (Bio-Rad, Hercules, CA, USA).
Pectin methylesterase enzyme inhibitor assay
The inhibitory effect of CaPMEI1 on the enzymatic activity of pectin methylesterase (PME) was assayed under standard conditions (Grsic-Rausch and Rausch 2004). The reaction mixture comprised 894 μL 0.4 mM NAD in 50 mM phosphate buffer (pH 7.5), 80 μL 5% (w/v) pectin (Sigma) in H2O, 8 μL formaldehyde dehydrogenase (0.35 U; Sigma) and 8 μL alcohol oxidase (1.0 U; Sigma). After mixing, the reaction was started with the addition of 10 μL (7.8 mU) PME from orange peel (Sigma). To analyze CaPMEI1 inhibition of PME, 1 μL inhibitor solution (0.5 mg CaPMEI1 fusion protein, 10 mM Tris buffer [pH 7.5], 0.1 M NaCl) was mixed with 10 μL PME (7.8 mU) and 0.5 μL 3 M K-acetate buffer (pH 5.3), followed by incubation for 15 min at room temperature. All reaction temperatures were maintained at 25°C. Reaction rates were recorded continuously at 340 nm using a DU650 UV-visible spectrophotometer (Beckman, Fullerton, CA, USA). PME–PMEI interactions were determined by measuring the rate of NADH formation per minute at pH 7.5 and 25°C.
In vitro antifungal activity
To determine the antifungal activity of CaPMEI1, we examined its effect on plant pathogenic fungi. Fusarium oxysporum f.sp. matthiole and Alternaria brassicicola were incubated on potato dextrose agar (PDA) at 28°C for 1–2 weeks; B. cinerea was incubated at 20°C for 1–2 weeks. Fungi were grown in 48-well plates (Cell Wells™, Corning Glass Works, Corning, NY, USA) containing sterile 4× potato dextrose broth (PDB, 100 μL) and a range of concentrations of purified CaPMEI1 protein (0–500 μg mL−1). Spore suspensions (104 spores mL−1) were prepared and 100 μL of inoculum was added to each microwell. Plates containing B. cinerea were incubated at 20°C; the other fungi were incubated at 28°C for 4–7 days.
To determine the inhibitory effect of CaPMEI1 on spore germination and hyphal growth, CaPMEI1 (0.1–500 μg mL−1) or the thioredoxin control were added to F. oxysporum f.sp matthiolae spore suspensions (105 spores mL−1), placed on glass slides and incubated for 12 h at 28°C. After 6 h incubation, 100 germinated spores were examined using a haemocytometer and the lengths of 50 individual hyphae were determined. The experiment was repeated three times.
Plasmid construction and plant transformation
The CaPMEI1 coding region was PCR-amplified without stop codon using the following primers to generate XbaI and BamHI sites: 5′-TCTAGAATGGAAGGTGGCA ATTTTCTCACA-3′ and 5′-GGATCCGCCGTGAAGGGCAGCCAGACGGT -3′. The fragment was inserted into pCR2.1-TOPO (Invitrogen) and digested with XbaI and BamHI. The construct was confirmed by sequencing. To generate the reporter construct p35S-CaPMEI1-GFP, the fragment was inserted into the XbaI–BamHI sites of the binary vector p35S-smGFP. p35S-smGFP was generated by fusing the gene encoding smGFP from the vector 326-GFP into pBIN35S digested with HindIII and EcoRI. The CaMV35S promoter-CaPMEI1-smGFP construct was introduced into the Agrobacterium tumefaciens strain EHA105 using electroporation.
Transgenic Arabidopsis plants were generated by floral-dipping wild-type (Col-0) plants into an A. tumefaciens culture containing the appropriate construct (Clough and Bent 1998). Agrobacterium was grown at 28°C and 250 rpm in YEP medium (10 g Bacto peptone, 10 g yeast extract, 5 g NaCl) supplemented with kanamycin (25 μg ml−1). Cells were harvested by centrifugation for 20 min at 5,500g and resuspended in inoculation media containing 5.0% sucrose and 0.05% Silwet L-77 (OSi Specialties, Inc., Danbury, CT, USA) to OD600 = 0.8. Arabidopsis plants with 2–10 cm long bolts were inoculated by dipping into an Agrobacterium suspension (OD600 = 0.8) and then left in the dark overnight, prior to return to a growth chamber. Seeds from the transformed Arabidopsis plants were collected and screened for kanamycin resistance.
Virus-induced gene silencing (VIGS)
The TRV-based VIGS system was used for silencing of CaPMEI1 in pepper plants, as described previously (Liu et al. 2002a; Chung et al. 2004). The pepper CaPMEI1 coding region was cloned into pTRV2 to generate the construct pTRV2:CaPMEI1. The fully extended cotyledons of 2-week-old pepper seedlings were co-infiltrated with A. tumefaciens GV3101 carrying pTRV1 or pTRV2:CaPMEI1 (OD600 = 0.2 for each construct). Plants were placed in a growth room at 25°C under a 16 h light/8 h dark photoperiod, to allow for growth and viral spread. Experiments were performed 5–6 weeks after the induction of silencing.
RT-PCR analysis
The RT reactions (20 μL) were performed at 42°C with total RNA (2 μg), oligo p(dT)15 primer (Roche, Mannheim, Germany) and AMV reverse transcriptase (Roche). Aliquots (1 μL) of the RT reaction products were used for RT-PCR analysis with the following gene-specific primers: 5′-CAGGATGCAACACTCTGGTGG-3′ and 5′-ATCAAAGGC CGGTTGGTC-3′ for CaBPR1 (accession no. AF053343); 5′-TGTCGAAGGTGGTCC AATAAA-3′ and 5′-TAGACAGAAGGATTGGCGAGG-3′ for CaPR10 (accession no. AF244121); 5′-ATCTGTACCAGCTTGCACGTGT-3′ and 5′-CCCTCACTGTGGCCT TGG-3′ for CaPOA1 (accession no. AF442387); and 5′-CAGGGAGATGAATTCTGA GGC-3′ and 5′-CATATGAACCTCTATGGATTTCTG-3′ for CaSAR82A (accession no. AF313766). RT-PCR conditions were 95°C for 10 min and 30 cycles of 95°C for 30 s, 52°C for 30 s, and 72°C for 1 min and 30 s. Single bands for PCR products were confirmed on an agarose gel.
Drought stress treatment and evaluation
For germination tests, seeds from Arabidopsis wild-type and CaPMEI1-OX transgenic lines were surface-sterilized and placed on MS media (Murashige and Skoog 1962) containing 200 or 600 mM mannitol. The seeds were maintained at 4°C for 48 h under dark conditions to synchronize germination, and then transferred to a growth chamber. Experiments were repeated at least three times, using approximately 100 seeds.
Surface-sterilized seeds were also used for examination of relative root length. Seedlings were grown on plates placed vertically in growth chambers and root length was estimated for 14 days. To assess mannitol tolerance, seedlings were grown for 7 days in 1× MS agar media supplemented with 1% sucrose, and then transferred to wells containing 1× MS liquid media supplemented with 100, 200, 300 or 400 mM mannitol. For drought treatment, 4-week-old soil-grown plants were deprived of water for 15 days and then re-watered on day 16. To minimize experimental variation, both the transgenic and control plants were grown in the same tray. Experiments were repeated at least three times.
For transpiration rate measurements, leaves of 4-week-old plants were detached and maintained at room temperature. Leaf weight was determined every 20 min for 2 h, and then every 1 h thereafter. Each measurement was performed using eight leaves. Experiments were repeated at least three times with similar results.
Oxidative stress treatment and evaluation
For analysis of oxidative stress tolerance, surface-sterilized seeds from Arabidopsis wild-type, vector control and CaPMEI1-OX transgenic lines were placed on MS medium containing different concentrations of methyl viologen (MV; Sigma). After synchronized germination, seeds were maintained in a growth chamber. To investigate whether or not MV treatment caused retardation of seedling development, 7-day-old seedlings were randomly transferred into well-plates containing 1× MS liquid medium supplemented with 0.05, 0.1 or 0.5 μM MV, and then grown for a further 2 weeks. To determine leaf senescence during MV treatment, fully expanded leaves were detached from 4-week-old plants and rinsed briefly with 70% ethanol. The detached leaves were floated on MS medium containing MV for 24 h following extraction with liquid nitrogen. Their chlorophyll content was measured spectrophotometrically according to the formula (Ca + b = 5.24 × A664 + 22.24 × A648), where C is the chlorophyll concentration (Ca + b) in micrograms per mL and A is absorption (Lichtenthaler 1987).
Results
Isolation and sequence analysis of CaPMEI1 cDNA in pepper
To identify the molecular mechanisms involved in plant defense against the invasion of microbial pathogens, we performed a differential hybridization screening of a cDNA library constructed from pepper leaves undergoing an incompatible interaction with Xcv (Jung and Hwang 2000). PR-1, PR-10, SAR8.2 and Myb transcription factor were among the pathogen-induced genes isolated by this screen. In addition, we isolated a full-length cDNA (834 bp) designated CaPMEI1 (C apsicum a nnuum pectin methylesterase inhibitor protein, accession no. AF477956), which contains a 54 bp 5′ untranslated sequence and a 174 bp 3′ untranslated region. The predicted open reading frame encodes a full-length protein of 200 amino acid residues, with an estimated molecular mass of 21.39 kDa and an isoelectric point (pI) of 6.51.
BLAST searches were performed between the predicted amino acid sequence of CaPMEI1S (accession no. AF477956) from C. annuum and the EMBL/GenBank database. These sequence analyses revealed that CaPMEI1 contains a conserved pectin methylesterase inhibitor (PMEI) domain of 156 amino acids (Fig. 1). Four cysteine residues were conserved in all the aligned proteins (Fig. 1), and these are predicted to be involved in the formation of disulfide bridges (Camardella et al. 2000). The predicted amino acid sequence of CaPMEI1 is 80% identical to that of Nicotiana tabacum DC1.2 protein (accession no. BAA95794) and has 50% identity with Arabidopsis thaliana ripening-related protein (accession no. BAA97200), 36% with Pinus radiata pectinesterase homolog (accession no. T08112) and 24% with A. thaliana invertase homolog (accession no. NP_201267).
Amino acid sequence alignments of pepper CaPMEI1 with Nicotiana tabacum DC1.2 protein (accession no. BAA95794), A. thaliana ripening-related protein (accession no. BAA97200), A. thaliana invertase homolog (accession no. NP201267) and Pinus radiata pectinesterase homolog (accession no. T08112). The boxed amino acid sequences represent the pectin methylesterase inhibitor (PMEI) domain. The conserved cysteine residues are marked by asterisks and the shaded regions represent conserved amino acid residues
Organ-specific expression of CaPMEI1 in pepper tissues
RNA gel blot analysis was performed to examine the organ-specific expression of CaPMEI1 in pepper plants. Although high levels of CaPMEI1 transcription were observed in stem tissues, only relatively low levels of expression were found in leaf, root, flower and green fruit (Fig. 2a). CaPMEI1 transcripts were not detected in red fruit.
RNA gel blot analysis of expression of CaPMEI1, CaSAR82A and CaBPR1 in pepper plants. Membranes were hybridized with probes from the 3′ UTR region of pepper CaPMEI1 cDNA or full-length CaBPR1 cDNA. Equal loading (20 μg) was verified by visualizing RNA on a gel stained with ethidium bromide. H healthy, M mock-inoculated or mock-treated. a CaPMEI1 expression in various organs of pepper plants. b Expression of CaPMEI1 and CaBPR1 in pepper leaves at various time intervals after inoculation with C. coccodes or virulent strain Ds1 (compatible interactions with pepper: susceptible response) and avirulent strain Bv5-4a (incompatible interactions with pepper: resistant response) of Xcv. c CaPMEI1 expression in lower (local) infected and upper (systemic) uninfected leaves at various time intervals after inoculation with the virulent and avirulent strains Ds1 and Bv5-4a of Xcv, P. fluorescence ATCC13525 and E. coli JM109. The lower leaves of pepper plants were inoculated at the 6-leaf stage. For the mock-inoculation, the lower leaves were infiltrated with 10 mM MgSO4. d Expression of CaPMEI1 and CABPR1 in pepper leaves at various time intervals after treatment with salicylic acid (SA, 5 mM), methyl jasmonate (MeJA, 100 μM), ethylene (5 μl L−1) and abscisic acid (100 μM). e Expression of CaPMEI1 and CaSAR82A in pepper leaves at various time intervals after treatment with drought, wounding, cold and H2O2
CaPMEI1 induction by pathogen infection
To assess the accumulation of CaPMEI1 transcripts in pepper leaves during the compatible and incompatible interactions with pathogens, leaves were inoculated with C. coccodes and Xcv Ds1 (virulent, compatible), as well as Xcv Bv5-4a (avirulent, incompatible). CaPMEI1 transcripts were not detected in leaves mock-inoculated with sterilized water (Fig. 2b, c). During fungal infection at the four- and eight-leaf stages, CaPMEI1 expression was induced at 48 and 12–48 h after inoculation, respectively, suggesting that the adult plant stage is more resistant to C. coccodes (Fig. 2b, upper panel). CaPMEI1 transcripts increased rapidly within 30 min after infection by both the virulent (Ds1) and avirulent (Bv5-4a) strains of Xcv (Fig. 2b, lower panel). High transcript levels accumulated by 18 h after infection with the virulent strain (Ds1) that causes a susceptible response; however, they started to decline rapidly by this time following infection with the avirulent strain (Bv5-4a) that causes a resistant response. CaBPR1 (Capsicum annuum basic PR1) expression was observed to confirm the success of individual inoculations. CaBPR1 transcripts were detected at 6 h after inoculation with the virulent and avirulent strains of Xcv and increased gradually during the 24 h following inoculation (Fig. 2b).
Local and systemic expression of CaPMEI1 in infected pepper plants
The local and systemic CaPMEI1 expression was analyzed in pepper leaves inoculated with Xcv and the non-pathogenic bacteria Pseudomonas fluorescens ATCC13525 and E. coli JM109 (Fig. 2c). In leaves infected with the virulent (Ds1) and avirulent (Bv5-4a) strains of Xcv, CaPMEI1 transcripts were detected 3 h after inoculation and levels decreased thereafter. Infection of lower (local) leaves by either Xcv strain induced systemic accumulation of CaPMEI1 transcripts in the upper leaves. The systemic induction by the avirulent strain (Bv5-4a) infection was stronger than that by the virulent strain (Ds1) infection. Local or systemic induction of CaPMEI1 transcripts was not observed 30 h after infection. In response to the non-pathogenic bacteria P. fluorescens and E. coli, CaPMEI1 transcripts were detected 3 h after inoculation and decreased subsequently in both local and systemic leaves.
CaPMEI1 induction following treatment with plant hormones
Plant hormones are involved in defense-related signal transduction pathways (Hammond-Kosack and Jones 1996; Chung et al. 2007). We examined whether or not CaPMEI1 expression is induced by plant hormones using pepper leaves treated with salicylic acid (SA), ethylene, methyl jasmonate (MeJA) or abscisic acid (ABA) (Fig. 2d). Salicylic acid strongly induced expression of CaPMEI1 at 1 and 2 h, as well as between 18 and 24 h after treatment. Following treatment with ethylene and MeJA, CaPMEI1 expression was strongly induced for 24 h. In addition, strong CaPMEI1 induction was observed 1 h after treatment with ABA.
In comparison, SA only induced CaBPR1 transcription 18–24 h after treatment (Fig. 2d), although transcripts were detected 6 h after treatment with ethylene and MeJA. In response to ethylene treatment, CaBPR1 transcripts increased rapidly between 18 and 24 h after treatment, but increased gradually between 6 and 24 h after MeJA treatment. CaBPR1 expression was induced between 2 and 12 h after ABA treatment.
CaPMEI1 induction by abiotic stresses
Abiotic stresses resulted in differential induction of CaPMEI1 in pepper leaves (Fig. 2e). The CaPMEI1 gene was responsive to mechanical injury, and expression was observed between 15 min and 6 h after wounding, although no transcription was detected thereafter. CaPMEI1 transcription also increased between 2 and 18 h after cold treatment. CaPMEI1 transcription was induced at 1 h of drought stress, but was undetectable thereafter. In response to H2O2 treatment, CaPMEI1 transcript levels increased for the first 6 h and declined thereafter. The marker genes CaBPR1 and CaSAR82A were used for comparison, as they also exhibit differential induction by these abiotic stresses.
In situ localization of CaPMEI1 transcripts
We performed an in situ hybridization using a CaPMEI1 probe to examine spatial expression of CaPMEI1 in pepper leaf tissues (Fig. 3). No hybridization signals were observed in non-treated leaves (Fig. 3a, b). Intense localization of CaPMEI1 transcripts was observed in the xylem area of the vascular bundle in non-treated stems (Fig. 3c). CaPMEI1 transcripts were detected in the vascular bundle of leaves inoculated with C. coccodes (Fig. 3e) and in the vascular bundles and upper epidermis of leaves treated with ethylene (Fig. 3g). No transcript signals were observed in any pepper tissues hybridized with the CaPMEI1 sense DIG-labeled RNA probe (Fig. 3b, d–f). These in situ hybridization results are supported by the temporal expression patterns observed in the RNA gel blot analysis of CaPMEI1 expression (Fig. 2).
In situ localization of CaPMEI1 transcripts in pepper leaf and stem tissues. Cross sections of leaf tissues were hybridized with CaPMEI1 antisense (a, c, e, g) and sense (b, d, f, h) DIG-labeled RNA probes, and then photographed under bright-field conditions. The transcript signal is purple. a, b Untreated leaf tissues. c, d Untreated stems. e, f Leaf tissues at 24 h after inoculation with C. coccodes. g, h Leaf tissues treated with 5 μl L−1 ethylene. P phloem, X xylem, UE upper epidermis, LE lower epidermis, Vs vascular bundle, C cortical cell
Inhibition of pectin methylesterase (PME) activity by CaPMEI1 protein
To prepare recombinant CaPMEI1 protein, the CaPMEI1 coding region was PCR-amplified and cloned into pET32a. E. coli strain BL21 (DE3) pLysS was used as a host for the recombinant expression of the CaPMEI1 construct and an empty vector control, yielding recombinant CaPMEI1 and thioredoxin, respectively. The purified CaPMEI1 (Fig. 4a, lane 5) and thioredoxin (Fig. 4a, lane 3) were examined by SDS-PAGE analysis. The purified thioredoxin–CaPMEI1 fusion protein formed a single band with an apparent molecular mass of 42 kDa (Fig. 4a, lane 5). Following cleavage of thioredoxin by enterokinase digestion, the purified CaPMEI1 protein (Fig. 4a, lane 6) was used for biological function determination. The cleaved recombinant CaPMEI1 protein was found exclusively in the insoluble fraction as inclusion bodies (data not shown). After attempting unsuccessfully to renature the cleaved protein using Triton X-100 as a detergent (Kim and Hwang 1994), we performed the PME inhibition assay using crude and purified CaPMEI1 fusion proteins.
Inhibition of pectin methylesterase (PME) activity by the pepper pectin methylesterase inhibitor (CaPMEI). a Recombinant CaPMEI1 expression in E. coli BL21 (DE3) pLysS. Cells were grown in LB media, and recombinant protein expression was induced with 1 mM IPTG. M, molecular marker (kDa); Lane 1, Uninduced E. coli BL21 cell extracts; Lane 2, Soluble fraction of E. coli BL21 cell extracts encoding thioredoxin protein after IPTG induction; Lane 3, Purified thioredoxin; Lane 4, Crude protein extracts of E. coli cells producing the thioredoxin–CaPMEI1 fusion protein after IPTG induction; Lane 5, Purified thioredoxin–CaPMEI1 fusion protein; Lane 6, Cleaved CaPMEI1 and thioredoxin proteins following enterokinase digestion. Protein staining was performed using Coomassie brilliant blue. b Inhibition of PME activity following treatment with CaPMEI1. For the inhibition assay, crude CaPMEI1 (0.5 μg) was mixed with 7.8 mU of orange peel PME in a total volume of 11 μL, and then preincubated at 25°C for 15 min, followed by addition to the reaction solution
CaPMEI1 contains a pectin methylesterase inhibitor (PMEI) domain (Fig. 1) and shares similarity with a plant pectin methylesterase inhibitor related to regulation of pectin degradation (Raiola et al. 2004). In addition, PMEIs have been found to play crucial roles in plant and microbial enzyme regulation of pectin degradation by de-esterification (Raiola et al. 2004). To determine whether or not orange peel PME activity is inhibited in the presence of crude CaPMEI1 fusion protein, we used the PME enzyme assay to measure the effect of PME inhibitor on the formation of NADH (Fig. 4b). Both crude and purified CaPMEI1 (ca. 0.5 μg) strongly inhibited PME enzyme activity, and a higher concentration of purified CaPMEI1 (ca. 1.0 μg) exhibited the strongest inhibition. However, thioredoxin alone did not exhibit any inhibitory effect. These results suggest that CaPMEI1 functions as an inhibitor of PME activity.
Antimicrobial activity of CaPMEI1
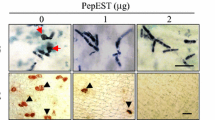
CaPMEI1 exhibited antifungal activity against the three plant pathogenic fungi examined: F. oxysporum f.sp. matthiole, A. brassicicola and B. cinerea (Fig. 5a). The thioredoxin–CaPMEI1 fusion protein (50 μg mL−1) suppressed mycelial growth of the three plant pathogenic fungi, whereas thioredoxin alone did not. Furthermore, the purified thioredoxin–CaPMEI1 (500 μg mL−1) fusion protein inhibited spore germination and hyphal growth of F. oxysporum f.sp. matthiole, whereas thioredoxin alone did not (Fig. 5b). Increasing concentrations of recombinant thioredoxin–CaPMEI1 fusion protein increasingly inhibited both spore germination and hyphal growth (Fig. 5c).
Assay of thioredoxin–CaPMEI1 fusion protein antimicrobial activity. a Inhibitory effects of the CaPMEI1–thioredoxin fusion protein on mycelial growth of the plant pathogenic fungi F. oxysporum f.sp. matthiolae, A. brassicicola and B. cinerea. In each plate, the upper wells (a) were treated with purified thioredoxin and the lower wells (b) were treated with the purified thioredoxin-CaPMEI1 fusion protein. b Inhibition of germination and hyphal growth of F. oxysporum f.sp. matthiolae. Fungal spores were allowed to germinate and grow in 100 μL potato dextrose broth medium alone (top), or with 500 μg mL−1 thioredoxin (middle) or 500 μg mL−1 thioredoxin–CaPMEI1 fusion protein (bottom). Photographs were taken after incubation for 10 h at 28°C. Bars 20 μm. c Inhibition of spore germination and hyphal growth of F. oxysporum f.sp. matthiolae by CaPMEI1. The percentage of germinated spores and the length of fungal hyphae were determined by light microscopy. Data represent means ± SD from three independent experiments
Enhanced susceptibility of CaPMEI1-silenced pepper plants to Xcv infection
Since inoculation with Xcv strongly induced CaPMEI1 expression in pepper plants, we performed the virus-induced gene silencing (VIGS) technique (Liu et al. 2002a; Chung et al. 2004) to examine its cellular function during pathogen infection. The full-length ORF of CaPMEI1 was used to construct pTRV2:CaPMEI1. Five to six weeks after induction of silencing, empty vector control (TRV:00) and CaPMEI1-silenced (TRV:CaPMEI1) pepper plants were inoculated with virulent and avirulent strains Ds1 and Bv5-4a of Xcv, respectively. To assess the efficiency of VIGS, CaPMEI1 transcript levels were examined by RT-PCR (Fig. 6). CaPMEI1 transcripts were nearly undetectable in both non-inoculated empty vector control and CaPMEI1-silenced plants. However, at 12 h after inoculation with virulent and avirulent Xcv, we observed strong induction of CaPMEI1 in the empty vector control plants, whereas only weak or undetectable transcript levels were found in CaPMEI1-silenced plants. These results confirm the effective silencing of the target gene in pepper plants.
RT-PCR analysis of expression of CaPMEI1 and several defense-related genes in empty vector control (TRV:00) and CaPMEI1 gene-silenced (TRV:CaPMEI1) pepper plants 12 h after inoculation with the virulent (Ds1; C, compatible) and avirulent (Bv5-4a; I, incompatible) strains of Xcv (5 × 106 cfu mL−1). 18S rRNA levels were visualized as a loading control. This experiment was repeated three times with similar results. H healthy leaves, CaBPR1 basic pathogenesis-related protein 1, CaPR10 putative ribonuclease-like protein, CaPOA1 ascorbate peroxidase 1, and CaSAR82A SAR8.2
To determine whether or not the expression of defense-related genes is affected by bacterial infection in the silenced plants, we used RT-PCR to analyze transcript levels of some defense-related genes (Fig. 6). In the empty vector control plants, both CaBPR1 (basic pathogenesis-related protein 1) and CaPR10 (putative ribonuclease-like protein) were strongly induced by virulent and avirulent Xcv infection. However, induction of these genes was slightly reduced in CaPMEI1-silenced plants infected with virulent Xcv, but was unaffected in plants infected with avirulent Xcv. Thus, CaPMEI1 gene silencing did not alter the induction of CaPOA1 (ascorbate peroxidase) or CaSAR82A (SAR 8.2) in response to Xcv infection.
To examine the function of the CaPMEI1 gene in basal defense or gene-for-gene resistance, empty vector control and CaPMEI1-silenced pepper plants were infected with virulent or avirulent Xcv (Fig. 7). CaPMEI1 gene silencing significantly increased susceptibility to virulent Xcv infection, but not to avirulent Xcv infection. The CaPMEI1-silenced pepper leaves exhibited more severe disease symptoms 5 days after virulent Xcv inoculation than did empty vector control plants, and these symptoms were accompanied by severe chlorosis and enlarged water-soaked lesions (Fig. 7a). However, we did not observe any phenotypical changes in the cell death response of CaPMEI1-silenced plants following avirulent Xcv inoculation (Fig. 7a). At 3 days after inoculation with virulent Xcv, bacterial growth was tenfold greater in CaPMEI1-silenced plants than in the empty vector controls (Fig. 7b). However, silencing of CaPMEI1 conferred a slightly enhanced susceptibility to avirulent Xcv. These findings suggest that CaPMEI1 may function in basal resistance of pepper plants against Xcv infection, rather than gene-for-gene resistance.
Enhanced disease susceptibility of CaPMEI1-silenced pepper plants to infection by the virulent Xcv strain Ds1, but not the avirulent Xcv strain Bv5-4a. a Disease symptoms developed on the leaves at different time points after inoculation with the virulent Xcv strain Ds1 (5 × 106 cfu mL−1) and the avirulent Xcv strain Bv5-4a (various bacterial concentrations). b Bacterial growth in leaves of empty vector control (TRV:00) or CaPMEI1-silenced (TRV:CaPMEI1) pepper plants at different time points after inoculation with the virulent Xcv strain Ds1 or the avirulent Xcv strain Bv5-4a (104 cfu mL−1). Data represent the mean ± SD from three independent experiments
Enhanced resistance of CaPMEI1-OX plants to Pst DC3000
The p35S-CaPMEI1-GFP and p35S-GFP overexpression (OX) plants were generated by transformation of A. thaliana using the floral dipping method (Clough and Bent 1998). Plants transformed with the empty vector p35S-GFP were used as a control. Seeds (T1) were collected from each transformed plant and screened for resistance to kanamycin. Northern blot analysis was performed on T1 plants exhibiting kanamycin resistance to determine the integrity of the inserted transgene. Three T2 plants showing strong CaPMEI1 expression were selected for analyses in planta (Fig. 8a).
Responses of wild-type (Col-0) Arabidopsis and CaPMEI1-OX transgenic plants to infection with P. syringae pv. tomato DC3000. a RNA gel blot analysis confirming CaPMEI1 overexpression (OX) in the transgenic Arabidopsis lines. Total RNA (10 μg) was loaded into each lane. The 3′ UTR region of pepper CaPMEI1 cDNA was used as a probe. b Growth of Pst DC3000 in the leaves of wild-type and transgenic plants. The mature leaves of the 6-week-old plants were infiltrated with a Pst Dc3000 suspension (105 cfu mL−1), and the degree of bacterial growth was rated at 0, 2 and 4 days after inoculation. c Disease symptoms on leaves of 6-week-old plants infiltrated with virulent Pst DC3000 (105 cfu mL−1). d Expression of pathogen-related (PR) genes in transgenic plants. Northern blot analyses were performed with 10 μg total RNA prepared from 5-week-old leaves of the wild-type (WT), vector control (smGFP) and transgenic (CaPMEI1::smGFP) plants. The samples were collected at 5, 15 and 25 h following pathogen infiltration with a suspension of the virulent strain Pst DC3000 (105 cfu mL−1)
To determine the contribution of CaPMEI1 to Arabidopsis resistance, wild-type and CaPMEI1-OX plants were inoculated with virulent P. syringae pv. tomato DC3000 (Pst; 105 cfu mL−1). Bacterial titers were determined 4 days after inoculation. CaPMEI1-OX plants exhibited much less growth of Pst bacteria than wild-type or empty vector control plants (Fig. 8b). In the transgenic plants, this reduced bacterial multiplication was most pronounced at 4 days after inoculation. At 6 days after inoculation, wild-type (Col-0) and empty vector control plants developed typical chlorotic symptoms in the infected leaves, whereas the transgenic plants displayed few disease symptoms (Fig. 8c).
Expression of defense-related genes in CaPMEI1-OX plants
To gain insight into the role played by CaPMEI1 in PR gene induction, we examined the expression patterns of the well-established marker genes AtPR1a, AtPR2 and AtPR5 in the defense responses of wild-type, vector control and the CaPMEI1-OX Arabidopsis plants infected with virulent Pst DC3000 (Fig. 8d). The expression of AtPR1a and AtPR2 was very similar in the CaPMEI1-OX, wild-type and vector control plants. AtPR5 expression was not induced in either un-inoculated or inoculated, wild-type or vector control plants. However, there was significant expression of AtPR5 in the transgenic CaPMEI1-OX plants at 15 and 25 h after Pst DC3000 infection. AtPR1a, AtPR2 and AtPR5 expression is known to be regulated via the SA-dependent pathway in Arabidopsis (Uknes et al. 1992), and in nahG plants, lack of SA reduces expression of these three PR genes during pathogenesis (Delany et al. 1994; Nawrath and Metraux 1999). In our experiments, AtPDF1.2 (defensin) transcripts were not detected in wild-type, vector control or CaPMEI1-OX plants (data not shown).
Responses of wild-type and CaPMEI1-OX plants to Hyaloperonospora parasitica
We examined whether or not ectopic CaPMEI1 expression in Arabidopsis plants affected resistance to the virulent biotrophic oomycte pathogen H. parasitica isolate Noco2 (Fig. 9). Over 100 seedlings of both wild-type (Col-0) and CaPMEI1-OX transgenic lines were inoculated with spores of H. parasitica isolate Noco2 (5 × 104 conidiosporangia mL−1). At 7 days after inoculation, over 50 plants in each line were sampled to estimate the disease and assess the degree of asexual sporulation, which was quantified and expressed as the mean number of sporangiophores per cotyledon.
Responses of wild-type (Col-0) Arabidopsis and CaPMEI1-OX transgenic plants to infection with H. parasitica isolate Noco2. a Disease symptoms and trypan blue-stained pathogen structures on 7-day-old cotyledons of wild-type and transgenic plants 7 days after inoculation; dpi days post-inoculation. Bars 0.5 mm. b Quantification of asexual sporangiophores per cotyledon for at least 50 cotyledons of wild-type and transgenic plants 7 days after inoculation. The average number of sporangiophores produced on the cotyledons of wild-type and transgenic lines are shown below each of the lines tested
As shown in Fig. 9a, the cotyledons of wild-type and transgenic seedlings responded to H. parasitica infection by stimulating similar levels of asexual parasite sporulation. Their trypan blue-stained cotyledons also exhibited abundant hyphae, conidiospores and oospores (Fig. 9a). Quantitative disease ratings are shown in Fig. 9b. Both the transgenic and the wild-type plants showed a high level of susceptibility to H. parasitica isolate Noco2, and similar levels of heavy asexual sporulation (>20 sporangiophores per cotyledon) were observed on both lines.
Drought tolerance of CaPMEI1-OX plants
To investigate a possible role for CaPMEI1 in the dehydration response, we tested seed germination and seedling growth under osmotic stress. The seeds of wild-type, vector control and CaPMEI1-OX plants were placed on MS media supplemented with various concentrations of mannitol. We observed no significant differences in seed germination between these lines (Fig. 10a). However, treatment with 200 and 600 mM mannitol strongly inhibited germination in the wild-type and vector control plants compared with the CaPMEI1-OX plants.
Transgenic Arabidopsis CaPMEI1-OX lines exhibit enhanced tolerance to drought stress. a Seed germination in wild-type, smGFP and transgenic plants on the MS media containing 0, 200 and 600 mM mannitol. The data represent the mean ± SD of 100 seeds for each line tested. b Relative root length of wild-type, smGFP and transgenic lines in MS agar medium containing different concentrations of mannitol. Three independent experiments were performed with 40 seedlings of both wild-type and transgenic lines. c Drought tolerance test of transgenic seedlings. Wild-type, smGFP and transgenic lines were germinated and grown in 1× MS agar medium. Each seedling was transferred to liquid medium containing 100 mM mannitol. d Wild-type, smGFP and the CaPMEI1 transgenic Arabidopsis plants after 15 days without water. e Water loss from the excised leaves of wild-type, smGFP and transgenic plants. Data represent the mean ± SD from three independent experiments
We also tested the sensitivity of root growth to osmotic stress (Fig. 10b). Drought tolerance was observed during post-germination growth. The root growth of wild-type and vector control seedlings was inhibited in the presence of 150 and 200 mM mannitol. However, root elongation of CaPMEI1-OX plants was less sensitive to mannitol-induced osmotic stress and in comparison with wild-type and vector control plants, mutant seedlings grew well in liquid medium supplemented with 100 mM mannitol (Fig. 10c).
In addition, we noticed that adult CaPMEI1-OX plants exhibited enhanced drought tolerance. Following 14 days without water, wild-type and vector plants had withered severely, whereas the CaPMEI1-OX plants remained healthy. To determine the effect of CaPMEI1 on survival, these plants were rewatered on day 16; wild-type and vector control plants died, whereas the transgenic lines survived (Fig. 10d). Next, we examined transpiration rates by measuring fresh weight loss in detached leaves. The leaves of CaPMEI1-OX lines exhibited slightly slower water loss than those of wild-type or vector control plants (Fig. 10e). Together, these results indicate that CaPMEI1 overexpression enhanced water stress resistance.
Oxidative tolerance of CaPMEI1-OX plants
To investigate the response of CaPMEI1-OX plants to oxidative stresses, seeds of wild-type, vector control and CaPMEI1-OX plants were exposed to MS medium containing methyl viologen (MV) (Fig. 11). Treatment with 5 or 10 μM MV significantly inhibited germination of wild-type and vector control seeds compared with CaPMEI1-OX seeds (Fig. 11a). Seven-day-old seedlings were transferred to a medium containing different MV concentrations (0–0.5 μM) and grown for 2 weeks. Wild-type and vector control seedlings turned white and started to die after 15 days of MV treatment, but CaPMEI1-OX lines were much less affected by the treatment (Fig. 11b). Furthermore, these differences in plant phenotype were also observed with respect to the higher fresh weight of transgenic seedlings compared to those of wild-type and vector control plants (Fig. 11c). Detached leaves of 4-week-old plants were treated with 10 μM MV for 24 h, after which chlorophyll content was measured (Fig. 11d). The leaves of transgenic lines treated with MV retained more chlorophyll than those of wild-type or vector control plants.
Transgenic Arabidopsis CaPMEI1-OX lines exhibit tolerance to oxidative stress. a Effects of methyl viologen on the seed germination of transgenic lines. Seeds from wild-type, vector control and transgenic lines were plated on media with or without methyl viologen (MV, 5 and 10 μM) and incubated for 3 days. The data represent mean ± SD of 100 seeds for each line tested. b Phenotypes of wild-type, vector control and transgenic line seedlings treated with different concentrations of MV. c Fresh weights of seedlings grown in the indicated concentrations of MV for 2 weeks. The results are presented as the average fresh weight per seedling. Data represent mean ± SD from three independent experiments. d Chlorophyll content of MV-treated leaves of wild-type, vector control and transgenic plants, which were floated on 0, 0.05, 0.1 and 0.5 μM MV in MS medium and then incubated for 24 h in a growth chamber
Discussion
Plants possess a diverse range of cell wall-modified enzymes, which are post-transcriptionally regulated by numerous inhibitor-related proteins (Tymowska-Lalanne and Kreis 1998; Raush and Greiner 2004). Several invertase inhibitor-related proteins have been isolated from higher plants such as kiwi, tobacco and Arabidopsis (Greiner et al. 1998; Wolf et al. 2003; Giovane et al. 2004; Raiola et al. 2004). Attempts to characterize the activity of invertase inhibitor proteins from plant species other than kiwi or Arabidopsis have been either unsuccessful or resulted in the isolation of invertase inhibitors that share structural similarities with pectin methylesterase inhibitor proteins (PMEI), but which represent completely different target enzymes (Greiner et al. 1998; Scognamiglio et al. 2003). In this study, we identified and functionally characterized a novel pepper CaPMEI1 gene encoding a PMEI. This CaPMEI1 protein contains the four cysteine residues that are conserved among other PMEI proteins (Camardella et al. 2000; Sato et al. 2000). These residues are expected to be engaged in two disulfide bridges, which constitute a common structural motif within the PMEI domain (Camardella et al. 2000).
At the molecular level, CaPMEI1 expression was induced in pepper leaves by infection with bacterial pathogens and treatment with plant hormones such as SA, ethylene, MeJA and ABA. In particular, these hormone treatments strongly induced CaPMEI1 transcription, suggesting that this gene may be involved in the early stages of the active defense responses to bacterial pathogen infection and exogenous treatment with plant hormones.
In enzymatic assays, purified CaPMEI1 proteins significantly inhibited activity of plant pectin methylesterase (PME). In addition, CaPMEI1 exhibited antifungal activity against a broad range of plant pathogenic fungi, including F. oxysporum f.sp. matthiolae, A. brassicicola and B. cinerea. To penetrate the cuticular layer, plant fungal pathogens produce plant cell wall-degrading enzymes such as polygalacturonase, pectin lyase and cellulase (Collmer and Keen 1986). An aggressive Phaeosphaeria nodorum isolate was shown to produce high amounts of xylanase, cellulase, polygalacturonase and butyrate esterase in vitro (Lalaoui et al. 2000). Recently, it was found that plant PMEIs do not inhibit the PMEs produced by plant pathogens (Giovane et al. 2004; Di Matteo et al. 2005). However, Arabidopsis plants expressing either AtPMEI-1 or AtPMEI-2 showed reduced infection by B. cinerea (Lionetti et al. 2007). This finding suggests that the increased level of pectin methylesterification caused by overexpression of AtPMEI-1 results in the inhibition of fungal endopolygalacturonase activity. Thus, the increase in PMEI activity resulted in reduced accessibility for fungal pectin degrading-enzymes and hence provided increased resistance to pathogens (Boudart et al. 1998; Lionetti et al. 2007). Consistent with these findings, our results suggest that CaPMEI1 may function as part of a new group of plant pectin methylesterase inhibitors, which restrict fungal pathogen infection in plants.
We used virus-induced gene silencing (VIGS) to investigate the effect of CaPMEI1 loss-of-function in pepper plants during Xcv infection. The CaPMEI1-silenced plants were susceptible to Xcv infection, and in particular to infection with the virulent strain, which resulted in enhanced bacterial growth and reduced PR1 and PR10 gene expression. Basal resistance is activated during the compatible bacterial interaction which restricts the spread of pathogens in the host plants to a certain extent (Glazebrook 2001). In addition, basal resistance is also effective in retarding proliferation of a wide range of microbial pathogens (Chisholm et al. 2006), but it is dependent upon SA accumulation (Cao et al. 1994, 1997; Kinkema et al. 2000). Therefore, we conclude that CaPMEI1 expression may be involved in basal resistance by triggering downstream PR gene induction in pepper plants.
To determine the effect of CaPMEI1 gain-of-function in planta, we generated the CaPMEI1-OX Arabidopsis transgenic lines and investigated their response to P. syringae and H. parasitica infection, because these well-known model pathogens have been used extensively for the study of disease resistance mechanisms in Arabidopsis (Quirino and Bent 2003; Slusarenko and Schlaich 2003). The CaPMEI1-OX lines were resistant to Pst DC3000 infection, but not to infection by the biotrophic oomycete H. parasitica, which uses living cells as a nutrient source during the infection cycle (Alfano and Collmer 1996; Heath 2002). Moreover, as CaPMEI1 transcripts localize intensively in the xylem of vascular bundles in leaf tissues, they may not affect the accessibility of host plant cells to H. parasitica. In contrast, we hypothesize that intercellular growth of P. syringae may be restricted by the extracellular secretion of CaPMEI1 into host cells, resulting in triggering of the basal resistance response.
We found that fungal growth was inhibited by treatment with the recombinant CaPMEI1 protein in vitro, suggesting that CaPMEI1 may interfere directly with pathogen infection of host plants. Plant pathogenic microorganisms have been shown to produce a variety of pectinolytic enzymes that macerate and kill plant tissues (Collmer and Keen 1986). Cell wall fragments released by these pectinolytic enzymes may elicit the plant defense response (D’Ovidio et al. 2004). The polygalacturonase-inhibiting protein (PGIP) plays an important role in the recognition and inhibition of fungal polygalacturonase (PG). Overexpression of PGIPs in Arabidopsis not only significantly reduces disease symptoms, but also enhances defense gene activation during pathogen infection (Ferrari et al. 2003). Thus, we suggest that CaPMEI1 can disrupt invading pathogenic microorganisms by inhibiting pectin methylesterases produced by these pathogens.
The reduced bacterial growth observed in CaPMEI1-OX Arabidopsis lines may result from the expression of SA-inducible genes and CaPMEI1 overexpression. The SA-inducible genes such as PR1, PR2 and PR5 are activated in the SA defense pathway (Uknes et al. 1992). PR5 proteins are similar to thaumatin, which is a sweet-tasting protein from Thaumatococcus daniellii (Hu and Reddy 1997), and several pathogens can induce these proteins in a wide range of plant species (Ward et al. 1991; Hu and Reddy 1997; Reuber et al. 1998). Members of the PR5 group have been shown to exhibit antifungal activity against a broad spectrum of fungal pathogens (Coca et al. 2000) and to participate in the coordinated induction of systemic acquired resistance (SAR) against TMV (Ward et al. 1991). Therefore, the concomitant induction of PR genes may contribute to the enhanced resistance of CaPMEI1-OX Arabidopsis plants to bacterial pathogens.
The CaPMEI1-OX Arabidopsis lines showed a strong tolerance to drought stress, and CaPMEI1 overexpression resulted in reduced transpiration and enhanced root elongation. In contrast, transgene overexpression did not cause any obvious phenotypic differences under optimal growing conditions. Dehydration factors such as a mannitol and polyethyleneglycol (PEG) have been used to evaluate the effects of decreased water availability and simulate drought conditions in wild-type and CaPMEI1-OX Arabidopsis plants (Gupta and Kaur 2005; Verslues et al. 2006). Other plant invertase inhibitor-related protein genes such as NtCIF and NtVIF are also strongly induced by treatment with PEG or ABA (Rausch and Greiner 2004). However, the molecular and genetic roles played by PMEIs in drought and osmotic stress tolerance remain poorly understood.
Since plant responses to different abiotic stresses may be related to the accumulation of ROS and the mechanisms for their detoxification, the role played by ROS in stress signaling has been studied extensively (Apel and Hirt 2004). Methyl viologen binds to the thylakoid membranes of chloroplasts and in the presence of light, transfers electrons to O2 in a chain reaction causing continuous formation of superoxide radicals and oxidative stress (Asada 1996). The Arabidopsis CaPMEI1-OX lines exhibited tolerance to oxidative stress, both during seed germination and seedling growth. This tolerance to oxidative stress may reduce the damage caused by other stresses via the antioxidizing system, which suggests that CaPMEI1 overexpression results in detoxification of endogenous superoxide.
Here, we have determined that CaPMEI1 from pepper plays a role as an antifungal protein and has an inhibitory effect on PME. Furthermore, we have shown that Arabidopsis CaPMEI1-OX lines are resistant to bacterial pathogens. In addition, they exhibit tolerance to drought and oxidative stress. In conclusion, these multivariate functions of CaPMEI1 provide valuable insights into understanding the physiological significance of PMEIs in plant disease resistance and abiotic stress tolerance.
References
Alfano JR, Collmer A (1996) Bacterial pathogens in plants: life up against the wall. Plant Cell 8:1683–1698
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55:373–399
Asada K (1996) Radical production and scavenging in the chloroplasts. In: Baker NR (ed) Photosynthesis and the environment. Kluwer, Netherlands, pp. 123–150
Asoufi H, Hameed KM, Mahasneh A (2007) The cellulase and pectinase activities associated with the virulence of indigenous Sclerotinia sclerotiorum isolates in Jordan Valley. Plant Pathol J 23:233–238
Balestrieri C, Castaldo D, Giovane A, Quagliuolo L, Servillo L (1990) A glycoprotein inhibitor of pectin methylesterase in kiwi fruit (Actinidia chinensis). Eur J Biochem 193:183–187
Boccara M, Chatain V (1989) Regulation and role in pathogenicity of Erwinia chrysanthemi. J Bacteriol 171:4085–4087
Boudart G, Lafitte C, Barthe JP, Frasez D, Esquerre-Tugaye MT (1998) Differential elicitation of defense responses by pectic fragments in bean seedlings. Planta 206:86–94
Bosch M, Cheung AY, Hepler PK (2005) Pectin methylesterase, a regulator of pollen tube growth. Pant Physiol 138:1334–1346
Brigneti G, Martin-Hernanez AM, Jin H, Chen J, Baulcombe DC, Baker B, Jones JDG (2004) Virus-induced gene silencing in Solanum species. Plant J 39:264–272
Camardella L, Carratore V, Ciardiello MA, Servillo L, Balestrieri C, Giovane A (2000) Kiwi protein inhibitor of pectin methylesterase. Eur J Biochem 267:4561–4565
Cao H, Bowling SA, Gordon S, Dong X (1994) Characterization of an Arabidopsis mutant that is non-responsive to inducers of systemic acquired resistance. Plant Cell 6:1583–1592
Cao H, Glazebrook J, Clark JD, Volko S, Dong X (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88:57–64
Chen MH, Sheng J, Hind G, Handa AK, Citovsky V (2000) Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for viral cell-to-cell movement. EMBO J 19:913–920
Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host–microbe interactions: shaping the evolution of the plant immune response. Cell 124:803–814
Chomczynski P, Sacchi N (1987) Single step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159
Chung E, Oh SK, Park JM, Choi D (2007) Expression and promoter analyses of pepper CaCDPK4 (Capsicum annuum calcium dependent protein kinase 4) during plant defense response to incompatible pathogen. Plant Pathol J 23:76–89
Chung E, Seong E, Kim YC, Chung EJ, Oh SK, Lee S, Park JM, Joung YH, Choi D (2004) A method of high frequency virus-induced gene silencing in chili pepper (Capsicum annuum L. cv. Bukang). Mol Cell 17:377–380
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Coca MA, Barbara D, Yun DJ, Hasegawa PM, Bressan RA, Narasimhan ML (2000) Heterotrimeric G-proteins of a filamentous fungus regulate cell wall composition and susceptibility to a plant PR-5 protein. Plant J 22:61–69
Collmer A, Keen NT (1986) The role of pectic enzymes in plant pathogenesis. Annu Rev Phytopathol 24:383–409
de Vries RP, Visser J (2001) Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol Mol Biol Rev 65:497–522
Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, Ryals J (1994) A central role of salicylic acid in plantresistance. Science 266:1247–1250
Denès JM, Baron A, Renard CM, Péan C, Drilleau JF (2000) Different action patterns for apple pectin methylesterase at pH 7.0 and 4.5. Carbohydr Res 327:385–393
Di Matteo A, Giovane A, Raiola A, Camardella L, Bonivento D, De Lorenzo G, Cervone F, Bellincampi D, Tsernoglou D (2005) Structural basis for the interaction between pectin methylesterase and a specific inhibitor protein. Plant Cell 17:849–858
D’Ovidio R, Mattei B, Roberti S, Bellincampi D (2004) Polygalacturonases, polygalacturonase-inhibiting proteins and pectic oligomers in plant–pathogen interactions. Biochim Biophys Acta 1696:237–244
Ferrari S, Vairo D, Ausubl FM, Cervone F, Lorenzo GD (2003) Tandemly duplicated Arabidopsis genes that encode polygalacturonase-inhibiting proteins are regulated coordinately by different signal transduction pathways in response to fungal infection. Plant Cell 15:93–106
Ferrari S, Galletti R, Vario D, Cervone F, De Lorenzo G (2006) Antisense expression of the Arabidopsis thaliana AtPGIP1 gene reduced polygalacturonase-inhibiting protein accumulation and enhances susceptibility to Botrytis cinerea. Mol Plant Microbe Interact 19:931–936
Giovane A, Servillo L, Balestrieri C, Raiola A, D’Avino R, Tamburrini M, Ciardiello MA, Camardella L (2004) Pectin methylesterase inhibitor. Biochim Biophys Acta 1696:245–252
Glazebrook J (2001) Genes controlling expression of defense responses in Arabidopsis—2001status. Curr Opin Plant Biol 4:301–308
Greiner S, Krausgrill S, Rausch T (1998) Cloning of a tobacco apoplasmic invertase inhibitor. Proof of function of the recombinant protein and expression analysis during plant development. Plant Physiol 116:733–742
Greiner S, Rausch T, Sonnewald U, Herbers K (1999) Ectopic expression of a tobacco invertase inhibitor homolog prevents cold-induced sweetening of potato tubers. Nat Biotechnol 17:708–711
Grsic-Rausch S, Rausch T (2004) A coupled spectrophotometric enzyme assay for the determination of pectin methylesterase activity and its inhibition by proteinaceous inhibitors. Anal Biochem 333:14–18
Gupta AK, Kaur N (2005) Sugar signalling and gene expression in relation to carbohydrate metabolism under abiotic stresses in plants. J Biosci 30:761–776
Hagerman AE, Austin PJ (1986) Continuous spectrophotometric assay for plant pectin methyl esterase. J Agric Food Chem 34:440–444
Hammond-Kosack KE, Jones DG (1996) Resistance gene-dependent plant defense reponses. Plant Cell 8:1773–1791
Han CU, Lee CH, Choi GJ, Kim J, Ahn S, Choi JE, Cha JS, Cho KY, Lee SW (2005) Simultaneous expression of the protease inhibitors in a rice blast-resistant mutant. Plant Pathol J 21:402–405
Heath MC (2002) Cellular interactions between biotrophic fungal pathogens and host and nonhost plants. Can J Plant Pathol 24:259–264
Hothorn M, Wolf S, Aloy P, Greiner S, Scheffzek K (2004) Structural insights into the target specificity of plant invertase and pectin methylesterase inhibitory proteins. Plant Cell 16:3437–3447
Hu X, Reddy ASN (1997) Cloning and expression of a PR5-like protein from Arabidopsis: inhibition of fungal growth by bacterially expressed protein. Plant Mol Biol 34:949–959
Jing CM, Lai YJ, Chang WH, Wu MC, Chang HM (2001) Pectinesterase inhibitor in jelly fig (Ficus awkeotsang cv. Makino) achenes. J Food Sci 66:225–228
Jing CM, Li CP, Chang JC, Chang HM (2002) Characterization of pectinsterase inhibitor in jelly fig (Ficus awkeotsang cv. Makino) achenes. J Agric Food Chem 50:4890–4894
Jung HW, Hwang BK (2000) Isolation, partial sequencing, and expression of pathogenesis-related cDNA genes from pepper leaves infected by Xanthomonas campestris pv. vesicatoria. Mol Plant Microbe Interact 13:136–142
Kagan-Zur V, Tieman DM, Marlow SJ, Handa AK (1995) Differential regulation of polygalacturonase and pectin methylesterase gene expression during and after heat stress in ripening tomato (Lycopersicon esculentum Mill.) fruits. Plant Mol Biol 29:1101–1110
Kim YJ, Hwang BK (1994) Differential accumulation of β-1,3-glucanase and chitinase isoforms in pepper stems infected by compatible and incompatible isolates of Phytophthora capsici. Physiol Mol Plant Pathol 45:195–209
Kim YJ, Hwang BK (2000) Pepper gene encoding a basic pathogenesis-related 1 protein is pathogen and ethylene inducible. Physiol Plant 108:51–60
Kinkema M, Fan W, Dong X (2000) Nuclear localization of NRP1 is required for activation of PR gene expression. Plant Cell 12:2339–2350
Koch KE (1996) Carbohydrate-modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol 47:509–540
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lalaoui F, Halama P, Dumortier V, Paul B (2000) Cell wall-degrading enzymes produced in vitro by isolates of Phaeosphaeria nodorum differing in aggressiveness. Plant Pathol 49:727–733
Lee SC, Hong JK, Kim YJ, Hwang BK (2000) Pepper gene encoding thionin is differentially induced by pathogens, ethylene and methyl jasmonate. Physiol Mol Plant Pathol 56:207–216
Lichtenthaler HK (1987) Chlorophylls and carotenoids: pigments or photosynthetic biomembranes. Methods Enzymol 148:350–382
Lievens S, Goormachtig S, Herman S, Holsters M (2002) Patterns of pectin methylesterase transcripts in developing stem nodules of Sesbania rostrata. Mol Plant Microbe Interact 15:164–168
Lionetti V, Raiola A, Camardella L, Giovane A, Obel N, Pauly M, Favaron F, Cervone F, Bellincampi D (2007) Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol 143:1871–1880
Liu Y, Schiff M, Dinesh-Kumar SP (2002a) Virus-induced gene silencing in tomato. Plant J 31:777–786
Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP (2002b) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30:415–429
Liu Y, Schiff M, Serino G, Deng X-W, Dinesh-Kumar SP (2002c) Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene-mediated resistance response to tobacco mosaic virus. Plant Cell 14:1483–1496
Ly-Nguyen B, Van Loey AM, Smout C, Verlent I, Duvetter T, Hendrickx ME (2004) Effect of intrinsic and extrinsic factors on the interaction of plant pectin methylesterase and its proteinaceous inhibitor from kiwi fruit. J Agric Food Chem 52:8144–8150
Micheli F (2001) Pectin methylesterases: cell wall enzymes with important roles in plant physiology. Trends Plant Sci 6:414–419
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15:473–497
Nawrath C, Metraux JP (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11:1393–1404
Peart JR, Cook G, Feys BJ, Parker JE, Baulcombe DC (2002a) An EDS1 orthologue is required for N-mediated resistance against tobacco mosaic virus. Plant J 29:569–579
Peart JR, Lu R, Sadanandom A, Malcuit I, Moffett P, Brice DC, Schauser L, Jaggard DA, Xiao S, Coleman MJ, Dow M, Jones JD, Shirasu K, Baulcombe DC (2002b) Ubiquitin ligase-associated protein SGT1 is required for host and non-host disease resistance in plants. Proc Natl Acad Sci USA 99:10865–10869
Pelloux J, Rustérucci C, Mellerowicz EJ (2007) New insights into pectin methylesterase structure and function. Trends Plant Sci 12:267–277
Quirino BF, Bent AF (2003) Deciphering host resistance and pathogen virulence: the Arabidopsis/Pseudomonas interaction as a model. Mol Plant Pathol 4:517–530
Raiola A, Camardella L, Giovane A, Mattei B, De Lorenzo G, Cervone F, Bellincampi D (2004) Two Arabidopsis thaliana genes encode functional pectin methyesterase inhibitors. FEBS Lett 557:199–203
Rausch T, Krausgrill S, Greiner S (1998) Invertase-inhibitor, Patent application, WO 98/04722
Raush T, Greiner S (2004) Plant protein inhibitors of invertases. Biochim Biophys Acta 1696:253–261
Ren C, Kermode AR (2000) An increase in pectin methyl esterase activity accompanies dormancy breakage and germination of yellow cedar seeds. Plant Physiol 124:231–242
Reuber TL, Plotnikova JM, Dewdney J, Rogers EE, Wood W, Ausbel FM (1998) Correction of defense gene induction defects with powdery mildew susceptibility in Arabidopsis enhanced disease susceptibility mutants. Plant J 16:473–485
Robertson D (2004) VIGS vectors for gene silencing: many targets, many tools. Annu Rev Plant Biol 55:495–519
Sato S, Nakamura Y, Kaneko T, Katoh T, Asamizu E, Kotani H, Tabata S (2000) Structural analysis of Arabidopsis thaliana chromosome 5. X. Sequence features of the regions of 3,076,755 bp covered by sixty P1 and TAC clones. DNA Res 7:31–63
Scognamiglio MA, Ciardiello MA, Tamburrini M, Carratore V, Rausch T, Camardella L (2003) The plant invertase inhibitor shares structural properties and disulfide bridges arrangement with the pectin methylesterase inhibitor. J Protein Chem 22:363–369
Slusarenko A, Schlaich N (2003) Downy mildew of Arabidopsis thaliana caused by Hyaloperonospora parasitica (formerly Peronospora parasitica). Mol Plant Pathol 4:159–170
Tieman DM, Handa AK (1994) Reduction in pectin methylesterase activity modifies tissue integrity and cation levels in ripening tomato (Lycopersicon esculentum Mill.) fruits. Plant Physiol 106:429–436
Tymowska-Lalanne Z, Kreis M (1998) Expression of Arabidopsis thaliana invertase gene family. Planta 207:259–265
Uknes S, Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J (1992) Acquired resistance in Arabidopsis. Plant Cell 4:645–656
Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 45:523–539
Ward ER, Uknes SJ, Williams SC, Dincher SS, Wiederhold DL, Alexander DC, Ahl-Goy P, Metraux JP, Ryals JA (1991) Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3:1085–1094
Wietholter N, Graessner B, Mierau M, Mort AJ, Moerschbacher BM (2003) Differences in the methyl ester distribution of homogalacturonans from near-isogenic wheat lines resistant and susceptible to the wheat stem rust fungus. Mol Plant Microbe Interact 10:945–952
Willats WG, McCartney L, Mackie W, Knox JP (2001a) Pectin: cell biology and prospects for functional analysis. Plant Mol Biol 47:9–27
Willats WG, Orfila C, Linberg G, Bucholt HC, Van Abeleek GJWM, Voragen AGJ, Marcus SE, Christensen TMIE, Mikkelsen JD, Murray BS, Knox JP (2001b) Modulation of the degree and pattern of methyl-esterification of pectic homogalacturonan in plant cell walls. Implications for pectin methyl esterase action, matrix properties, and cell adhesion. J Biol Chem 276:19404–19413
Wolf S, Grsic-Rausch S, Rausch T, Greiner S (2003) Identification of pollen-expressed pectin methylesterase inhibitors in Arabidopsis. FEBS Lett 555:551–555
Acknowledgments
We thank Dr. S.P. Dinesh-Kumar (Yale University) for the pTRV1 and pTRV2 vectors; Dr. U. Bonas (Martin-Luther-Universitaet) for Agrobacterium tumefaciens strain GV3101 and Dr. Jonathan DG Jones (Sainsbury Laboratory) for Hyaloperonospora parasitica isolate Noco2. This work was supported by a grant (CG1133) from the Crop Functional Genomics Center of the 21st Century, Frontier Research Program, funded by the Ministry of Science and Technology, Korea, and a grant (20070401034028) from the Biogreen21 Program, Rural Development Administration, Korea.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Soo Hyun An and Kee Hoon Sohn contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
An, S.H., Sohn, K.H., Choi, H.W. et al. Pepper pectin methylesterase inhibitor protein CaPMEI1 is required for antifungal activity, basal disease resistance and abiotic stress tolerance. Planta 228, 61–78 (2008). https://doi.org/10.1007/s00425-008-0719-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-008-0719-z