Abstract
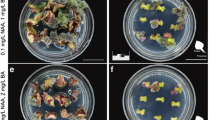
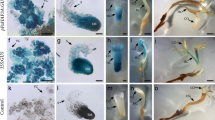
Lisianthus [Eustoma grandiflorum (Raf.) Shinn] is a popular cut flower crop throughout the world, and the demand for this plant for cut flowers and potted plants has been increasing worldwide. Recent advances in genetic engineering have enabled the transformation and regeneration of plants to become a powerful tool for improvement of lisianthus. We have established a highly efficient plant regeneration system and Agrobacterium-mediated genetic transformation of E. grandiflorum. The greatest shoot regeneration frequency and number of shoot buds per explant are observed on media supplemented with 6-Benzylaminopurine (BAP) and α-Naphthalene acetic acid (NAA). We report an efficient plant regeneration system using leaf explants via organogenesis with high efficiency of transgenic plants (15%) in culture of 11 weeks’ duration. Further ectopic expression of two MADS box genes, LMADS1-M from lily (Lilium longiflorum) and OMADS1 from orchid (Oncidium Gower Ramsey), was performed in E. grandiflorum. Conversion of second whorl petals into sepal-like structures and alteration of third whorl stamen formation were observed in the transgenic E. grandiflorum plants ectopically expressing 35S::LMADS1-M. 35S::OMADS1 transgenic E. grandiflorum plants flowered significantly earlier than non-transgenic plants. This is the first report on the ectopic expression of two MADS box genes in E. grandiflorum using a simple and highly efficient gene transfer protocol. Our results reveal the potential for floral modification in E. grandiflorum through genetic transformation.






Similar content being viewed by others
Abbreviations
- B5:
-
Gamborg
- BAP:
-
6-Benzylaminopurine
- CaMV:
-
Cauliflower mosaic virus
- CIM:
-
Callus induction medium
- IAA:
-
Indole-3-acetic acid
- MS:
-
Murashige and Skoog
- NAA:
-
α-Naphthalene acetic acid
- PCR:
-
Polymerase chain reaction
- RT-PCR:
-
Reverse transcription-PCR
- RIM:
-
Root induction medium
- SIM:
-
Shoot induction medium
References
Angenent GC, Busscher M, Franken J, Mol JNM, van Tunen AJ (1992) Differential expression of two MADS box genes in wild-type and mutant petunia flowers. Plant Cell 4:983–993
Angenent GC, Franken J, Busscher M, Weiss D, van Tunen AJ (1994) Co-suppression of the petunia homeotic gene fbp2 affects the identity of the generative meristem. Plant J 5:33–44
Aranovich D, Lewinsohn E, Zaccai M (2007) Post-harvest enhancement of aroma in transgenic lisianthus (Eustoma grandiflorum) using the Clarkia breweri benzyl alcohol acetyltransferase (BEAT) gene. Postharvest Biol Technol 43(2):255–260
Clark DG (2004) Applications of plant biotechnology to ornamental crops. In: Christou P, Klee H (eds) Handbook of plant biotechnology, vol 2: applications of plant biotechnology in agriculture, the pharmaceutical industry, other industries. Wiley, London, pp 863–879
Curtis IS, Power JB, Hedden P, Ward DA, Phillips A, Lowe KC, Davey MR (1999) A stable transformation system for the ornamental plant, Datura meteloides D.C. Plant Cell Rep 18:554–560
Favaro R, Pinyopich A, Battaglia R, Kooiker M, Borghi L, Ditta G, Yanofsky MF, Kater MM, Colombo L (2003) MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 15:2603–2611
Furukawa H, Matsubara C, Shigematsu N (1990) Shoot regeneration from the roots of prairie gentian [Eustoma grandiflorum (Griseb.) Schinners]. Plant Tissue Cult Lett 7:11–13
Gamborg O, Miller R, Ojima K (1968) Nutrients requirements of suspension cultures of soybean root cells. Exp Cell Res 50:151–158
Handa T (1992) Regeneration and characterization of prairie gentian (Eustoma grandiflorum) plant transformed by Agrobacterium rhizogenes. Plant Tissue Cult Lett 9:10–14
Handa T (1996) Transformation of prairie gentian (Eustoma grandiflorum) with Agrobacterium rhizogenes harboring GUS and NPTII genes. J Jpn Soc Hort Sci 64(4):913–918
Hsu HF, Huang CH, Chou LH, Yang CH (2003) Ectopic expression of an orchid (Oncidium Gower Ramsey) AGL6-like gene promotes flowering by activating flowering time genes in Arabidopsis thaliana. Plant Cell Physiol 44:783–794
Jack T, Brockman LL, Meyerowitz EM (1992) The homeotic gene APETALAE3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68:683–697
Kondo T, Hasegawa H, Suzuki M (2000) Transformation and regeneration of garlic (Allium sativum L.) by Agrobacterium mediated gene transfer. Plant Cell Rep 19:989–993
Kunitake H, Nakashima T, Mori K, Tanaka M, Mii M (1995) Plant regeneration from mesophyll protoplasts of lisianthus (Eustoma grandiflorum) by adding activated charcoal into protoplast culture medium. Plant Cell Tissue Organ Cult 43:59–65
Ledger SE, Deroles SC, Manson DG, Marie Bradley J, Given NK (1997) Transformation of lisianthus (Eustoma grandiflorum). Plant Cell Rep 16:853–858
Mandel MA, Yanofsky MF (1995) A gene triggering flower formation in Arabidopsis. Nature 377:522–524
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:475–497
Paek KY, Hahn EJ (2000) Cytokinins, auxins and activated charcoal affect organogenesis and anatomical characteristics of shoot-tip cultures of lisianthus (Eustoma grandiflorum (Raf.) Shinn). In Vitro Cell Dev Biol Plant 36:128–132
Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405:200–203
Pnueli L, Hareven D, Rounsley SD, Yanofsky MF, Lifschitz E (1994) Isolation of tomato AGAMOUS gene TAG1 and analysis of its homeotic role in transgenic plants. Plant Cell 6:163–173
Roh SM, Lawson RH (1988) Tissue culture in the improvement of Eustoma. Hortic Sci 23:658
Rounsley SD, Ditta GS, Yanofsky MF (1995) Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 7:1259–1269
Ruffoni B, Damiano C, Massabo F, Esposito P (1990) Organogenesis and embryogenesis in Lisianthus russellianus Hook. Acta Hortic 280:83–87
Semeria L, Vaira AM, Accotto GP, Allavena A (1995) Genetic transformation of Eustoma grandiflorum Griseb by microprojectile bombardment. Euphytica 85:125–130
Semeria L, Ruffoni B, Rabaglio M, Genga A, Vaira AM, Accotto GP, Allavena A (1996) Genetic transformation of Eustoma grandiflorum by Agrobacterium tumefaciens. Plant Cell Tissue Organ Cult 47:67–72
Shitsukawa N, Tahira C, Kassai K, Hirabayashi C, Shimizu T, Takumi S, Mochida K, Kawaura K, Ogihara Y, Murai K (2007) Genetic and epigenetic alteration among three homoeologous genes of a class E MADS box gene in hexaploid wheat. Plant Cell 19:1723–1737
Takahashi M, Nishihara M, Yamamura S, Nishizawa S, Irifune K, Morikawa H (1998) Stable transformation of Eustoma grandiflorum by particle bombardment. Plant Cell Rep 17:504–507
Theissen G (2001) Development of floral organ identity: stories from the MADS house. Curr Opin Plant Biol 4:75–85
Theissen G, Saedler H (2001) Floral quartets. Nature 409:469–471
Tzeng TY, Yang CH (2001) A MADS box gene from lily (Lilium longiflorum) is sufficient to generate dominant negative mutation by interacting with PISTILLATA (PI) in Arabidopsis thaliana. Plant Cell Physiol 42(10):1156–1168
Tzeng TY, Hsiao CC, Chi PJ, Yang CH (2003) Two lily SEPALLATA-like genes cause different effects on floral formation and floral transition in Arabidopsis thaliana. Plant Physiol 133:1091–1101
Weigel D, Meyerowitz EM (1994) The ABCs of floral homeotic genes. Cell 78:203–209
Zahn LM, Kong H, Leebens-Mack JH, Kim S, Soltis PS, Landherr LL, Soltis DE, dePamphilis CW, Ma H (2005) The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics 169:2209–2223
Acknowledgments
This work was supported by grants to C.-H. Y from the Council of Agriculture, Taiwan, ROC, grant number: 97AS-1.1.1-FD-Z1. This work was also supported by a 5Y/50B grant from the Ministry of Education.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Jordan.
Rights and permissions
About this article
Cite this article
Thiruvengadam, M., Yang, CH. Ectopic expression of two MADS box genes from orchid (Oncidium Gower Ramsey) and lily (Lilium longiflorum) alters flower transition and formation in Eustoma grandiflorum . Plant Cell Rep 28, 1463–1473 (2009). https://doi.org/10.1007/s00299-009-0746-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-009-0746-7