Abstract
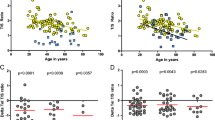
Complex karyotypes are associated with a poor prognosis in chronic lymphocytic leukemia (CLL). Using mFISH, iFISH, and T/C-FISH, we thoroughly characterized 59 CLL patients regarding parameters known to be involved in chromosomal instability: status of the genes ATM and TP53 and telomere length. Interestingly, a deletion of the ATM locus in 11q, independent of the cytogenetic context, was associated with significantly diminished risk (p<0.05) of carrying a mutation in TP53. In patients with loss or mutation of TP53, chromosomal breakage occurred more frequently (p<0.01) in (near-) heterochromatic regions. Median telomere length in patients with complex karyotypes was significantly shorter than that of healthy controls and shorter than in all other cytogenetic cohorts. Furthermore, the median telomere length of patients carrying a TP53 mutation was significantly shorter than without mutation. We conclude that telomere shortening in combination with loss of TP53 induces increased chromosomal instability with preferential involvement of (near-) heterochromatic regions.

Similar content being viewed by others
References
Dohner H, Stilgenbauer S, Benner A et al (2000) Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med 343:1910–1916
Hallek M (2013) Chronic lymphocytic leukemia: 2013 update on diagnosis, risk stratification and treatment. Am J Hematol 88:803–816
Rigolin GM, Del Giudice I, Formigaro L et al (2015) Chromosome aberrations detected by conventional karyotyping using novel mitogens in chronic lymphocytic leukemia: clinical and biologic correlations. Genes Chromosomes Cancer 54:818–826
Dohner H, Stilgenbauer S, James MR et al (1997) 11q deletions identify a new subset of B cell chronic lymphocytic leukemia characterized by extensive nodal involvement and inferior prognosis. Blood 89:2516–2522
Hallek M (2013) Signaling the end of chronic lymphocytic leukemia: new frontline treatment strategies. Blood 122:3723–3734
Dos Santos P, Panero J, Palau Nagore V, Stanganelli C, Bezares RF, Slavutsky I (2015) Telomere shortening associated with increased genomic complexity in chronic lymphocytic leukemia. Tumour Biol
Hoxha M, Fabris S, Agnelli L et al (2014) Relevance of telomere/telomerase system impairment in early stage chronic lymphocytic leukemia. Genes Chromosomes Cancer 53:612–621
Lin TT, Norris K, Heppel NH et al (2014) Telomere dysfunction accurately predicts clinical outcome in chronic lymphocytic leukaemia, even in patients with early stage disease. Br J Haematol 167:214–223
Zenz T, Mertens D, Dohner H, Stilgenbauer S (2011) Importance of genetics in chronic lymphocytic leukemia. Blood Rev 25:131–137
Veronese L, Tournilhac O, Callanan M et al (2013) Telomeres and chromosomal instability in chronic lymphocytic leukemia. Leukemia 27:490–493
Shay JW, Wright WE (2005) Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis 26:867–874
Rossi D, Gaidano G (2012) ATM and chronic lymphocytic leukemia: mutations, and not only deletions, matter. Haematologica 97:5–8
Lange K, Gadzicki D, Schlegelberger B, Gohring G (2010) Recurrent involvement of heterochromatic regions in multiple myeloma—a multicolor FISH study. Leuk Res 34:1002–1006
Hiller B, Bradtke J, Balz H, Rieder H (2005) CyDAS: a cytogenetic data analysis system. Bioinformatics 21:1282–1283
Gohring G, Karow A, Steinemann D et al (2007) Chromosomal aberrations in congenital bone marrow failure disorders—an early indicator for leukemogenesis? Ann Hematol 86:733–739
Heerema NA, Byrd JC, Dal Cin PS et al (2010) Stimulation of chronic lymphocytic leukemia cells with CpG oligodeoxynucleotide gives consistent karyotypic results among laboratories: a CLL research consortium (CRC) study. Cancer Genet Cytogenet 203:134–140
Perner S, Bruderlein S, Hasel C et al (2003) Quantifying telomere lengths of human individual chromosome arms by centromere-calibrated fluorescence in situ hybridization and digital imaging. Am J Pathol 163:1751–1756
Feurstein S, Rucker FG, Bullinger L et al (2014) Haploinsufficiency of ETV6 and CDKN1B in patients with acute myeloid leukemia and complex karyotype. BMC Genomics 15:784
Haferlach C, Dicker F, Schnittger S, Kern W, Haferlach T (2007) Comprehensive genetic characterization of CLL: a study on 506 cases analysed with chromosome banding analysis, interphase FISH, IgV (H) status and immunophenotyping. Leukemia 21:2442–2451
Thompson PA, O'Brien SM, Wierda WG et al (2015) Complex karyotype is a stronger predictor than del (17p) for an inferior outcome in relapsed or refractory chronic lymphocytic leukemia patients treated with ibrutinib-based regimens. Cancer 121:3612–3621
Skowronska A, Austen B, Powell JE et al (2012) ATM germline heterozygosity does not play a role in chronic lymphocytic leukemia initiation but influences rapid disease progression through loss of the remaining ATM allele. Haematologica 97:142–146
Bargetzi MJ, Muhlematter D, Tichelli A, Jotterand M, Wernli M (1999) Dicentric translocation (9;12) presenting as refractory Philadelphia chromosome-positive acute B cell lymphoblastic leukemia. Cancer Genet Cytogenet 113:90–92
Peters AH, O'Carroll D, Scherthan H et al (2001) Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107:323–337
Schotta G, Lachner M, Sarma K et al (2004) A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev 18:1251–1262
Stewart SA, Weinberg RA (2006) Telomeres: cancer to human aging. Annu Rev Cell Dev Biol 22:531–557
Blackburn EH (2001) Switching and signaling at the telomere. Cell 106:661–673
Blasco MA (2007) Telomere length, stem cells and aging. Nat Chem Biol 3:640–649
Alhourani E, Othman MA, Melo JB et al (2016) BIRC3 alterations in chronic and B-cell acute lymphocytic leukemia patients. Oncol Lett 11:3240–3246
Cortese D, Sutton LA, Cahill N et al (2014) On the way towards a “CLL prognostic index”: focus on TP53, BIRC3, SF3B1, NOTCH1 and MYD88 in a population-based cohort. Leukemia 28:710–713
Nadeu F, Delgado J, Royo C et al (2016) Clinical impact of clonal and subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM mutations in chronic lymphocytic leukemia. Blood 127:2122–2130
Foa R, Del Giudice I, Guarini A, Rossi D, Gaidano G (2013) Clinical implications of the molecular genetics of chronic lymphocytic leukemia. Haematologica 98:675–685
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
For this type of study formal consent is not required.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Funding
This study was funded by the Cluster of Excellence REBIRTH (from Regenerative Biology to Reconstructive Therapy, EXC 6273).
Additional information
Kathrin Thomay and Caroline Fedder shared first authorship
Rights and permissions
About this article
Cite this article
Thomay, K., Fedder, C., Hofmann, W. et al. Telomere shortening, TP53 mutations and deletions in chronic lymphocytic leukemia result in increased chromosomal instability and breakpoint clustering in heterochromatic regions. Ann Hematol 96, 1493–1500 (2017). https://doi.org/10.1007/s00277-017-3055-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00277-017-3055-1