Abstract
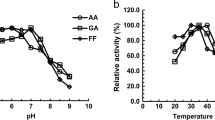
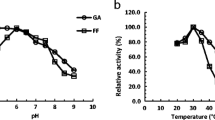
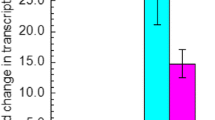
The short-chain dehydrogenase/reductase (SDR) family, the largest family in dehydrogenase/reductase superfamily, is divided into “classical,” “extended,” “intermediate,” “divergent,” “complex,” and “atypical” groups. Recently, several open reading frames (ORFs) were characterized as intermediate SDR aldehyde reductase genes in Saccharomyces cerevisiae. However, no functional protein in the atypical group has been characterized in S. cerevisiae till now. Herein, we report that an uncharacterized ORF YLL056C from S. cerevisiae was significantly upregulated under high furfural (2-furaldehyde) or 5-(hydroxymethyl)-2-furaldehyde concentrations, and transcription factors Yap1p, Hsf1p, Pdr1/3p, Yrr1p, and Stb5p likely controlled its upregulated transcription. This ORF indeed encoded a protein (Yll056cp), which was grouped into the atypical subgroup 7 in the SDR family and localized to the cytoplasm. Enzyme activity assays showed that Yll056cp is not a quinone or ketone reductase but an NADH-dependent aldehyde reductase, which can reduce at least seven aldehyde compounds. This enzyme showed the best Vmax, Kcat, and Kcat/Km to glycolaldehyde, but the highest affinity (Km) to formaldehyde. The optimum pH and temperature of this enzyme was pH 6.5 for reduction of glycolaldehyde, furfural, formaldehyde, butyraldehyde, and propylaldehyde, and 30 °C for reduction of formaldehyde or 35 °C for reduction of glycolaldehyde, furfural, butyraldehyde, and propylaldehyde. Temperature and pH affected stability of this enzyme and this influence varied with aldehyde substrate. Metal ions, salts, and chemical protective additives, especially at high concentrations, had different influence on enzyme activities for reduction of different aldehydes. This research provided guidelines for study of more uncharacterized atypical SDR enzymes from S. cerevisiae and other organisms.







Similar content being viewed by others
References
Buysschaert G, Verstraete K, Savvides SN, Vergauwen B (2013) Structural and biochemical characterization of an atypical short-chain dehydrogenase/reductase reveals an unusual cofactor preference. FEBS J 280(5):1358–1370. doi:10.1111/febs.12128
Drew D, Newstead S, Sonoda Y, Kim H, von Heijne G, Iwata S (2008) GFP-based optimization scheme for the overexpression and purification of eukaryotic membrane proteins in Saccharomyces cerevisiae. Nat Protoc 3(5):784–798. doi:10.1038/nprot.2008.44
Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Güldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kötter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418(6896):387–391. doi:10.1038/nature00935
Gietz RD, Schiestl RH, Willems AR, Woods RA (1995) Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11(4):355–360. doi:10.1002/yea.320110408
Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG (1996) Life with 6000 genes. Science 274(5287): 546, 563–567. doi:10.1126/science.274.5287.546
Heer D, Heine D, Sauer U (2009) Resistance of Saccharomyces cerevisiae to high concentrations of furfural is based on NADPH-dependent reduction by at least two oxireductases. Appl Environ Microbiol 75(24):7631–7638. doi:10.1128/AEM.01649-09
Iwahashi Y, Hosoda H, Park JH, Lee JH, Suzuki Y, Kitagawa E, Murata SM, Jwa NS, Gu MB, Iwahashi H (2006) Mechanisms of patulin toxicity under conditions that inhibit yeast growth. J Agric Food Chem 54(5):1936–1942. doi:10.1021/jf052264g
Jayakody LN, Hayashi N, Kitagaki H (2011) Identification of glycolaldehyde as the key inhibitor of bioethanol fermentation by yeast and genome-wide analysis of its toxicity. Biotechnol Lett 33(2):285–292. doi:10.1007/s10529-010-0437-z
Jörnvall H, Persson M, Jeffery J (1981) Alcohol and polyol dehydrogenases are both divided into two protein types, and structural properties cross-relate the different enzyme activities within each type. Proc Natl Acad Sci 78(7):4226–4230
Jörnvall H, Persson B, Krook M, Atrian S, Gonzalez-Duarte R, Jeffery J, Ghosh D (1995) Short-chain dehydrogenases/reductases (SDR). Biochemistry 34:6003–6013. doi:10.1021/bi00018a001
Jörnvall H, Höög JO, Persson B (1999) SDR and MDR: completed genome sequences show these protein families to be large, of old origin, and of complex nature. FEBS Lett 445(2–3):261–264. doi:10.1016/S0014-5793(99)00130-1
Jörnvall H, Landreh M, Östberg LJ (2015) Alcohol dehydrogenase, SDR and MDR structural stages, present update and altered era. Chem Biol Interact 234:75–79. doi:10.1016/j.cbi.2014.10.017
Kallberg Y, Persson B (2006) Prediction of coenzyme specificity in dehydrogenases/reductases: a hidden Markov model-based method and its application on complete genomes. FEBS J 273(6):1177–1184. doi:10.1111/j.1742-4658.2006.05153.x
Kallberg Y, Oppermann U, Jörnvall H, Persson B (2002) Short-chain dehydrogenases/reductases (SDRs): coenzyme-based functional assignments in completed genomes. Eur J Biochem 269(18):4409–4417. doi:10.1046/j.1432-1033.2002.03130.x
Kavanagh KL, Jörnvall H, Persson B, Oppermann U (2008) The SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell Mol Life Sci 65:3895–3906. doi:10.1007/s00018-008-8588-y
Kodo N, Matsuda T, Doi S, Munakata H (2013) Salicylic acid resistance is conferred by a novel YRR1 mutation in Saccharomyces cerevisiae. Biochem Biophys Res Commun 434(1):42–47. doi:10.1016/j.bbrc.2013.03.069
Larsson S, Palmqvist E, Hahn-Hägerdal B, Tengborg C, Stenberg K, Zacchi G, Nilvebrant NO (1999) The generation of inhibitors during dilute acid hydrolysis of softwood. Enzym Microb Technol 24:151–159. doi:10.1016/S0141-0229(98)00101-X
Li X, Yang R, Ma M, Wang X, Tang J, Zhao X, Zhang X (2015) A novel aldehyde reductase encoded by YML131W from Saccharomyces cerevisiae confers tolerance to furfural derived from lignocellulosic biomass conversion. Bioenerg Res 8(1):119–129. doi:10.1007/s12155-014-9506-9
Liu ZL (2011) Molecular mechanisms of yeast tolerance and in situ detoxification of lignocellulose hydrolysates. Appl Microbiol Biotechnol 90:809–825. doi:10.1007/s00253-011-3167-9
Liu ZL, Moon J (2009) A novel NADPH-dependent aldehyde reductase gene from Saccharomyces cerevisiae NRRL Y-12632 involved in the detoxification of aldehyde inhibitors derived from lignocellulosic biomass conversion. Gene 446(1):1–10. doi:10.1016/j.gene.2009.06.018
Liu ZL, Slininger PJ (2007) Universal external RNA controls for microbial gene expression analysis using microarray and qRT-PCR. J Microbiol Methods 68(3):486–496. doi:10.1016/j.mimet.2006.10.014
Liu ZL, Moon J, Andersh BJ, Slininger PJ, Weber S (2008) Multiple gene-mediated NAD(P)H-dependent aldehyde reduction is a mechanism of in situ detoxification of furfural and 5-hydroxymethylfurfural by Saccharomyces cerevisiae. Appl Microbiol Biotechnol 81(4):743–753. doi:10.1007/s00253-008-1702-0
Liu ZL, Ma M, Song M (2009a) Evolutionarily engineered ethanologenic yeast detoxifies lignocellulosic biomass conversion inhibitors by reprogrammed pathways. Mol Gen Genomics 282(3):233–244. doi:10.1007/s00438-009-0461-7
Liu ZL, Palmquist DE, Ma M, Liu J, Alexander NJ (2009b) Application of a master equation for quantitative mRNA analysis using qRT-PCR. J Biotechnol 143(1):10–16. doi:10.1016/j.jbiotec.2009.06.006
Lucau-Danila A, Delaveau T, Lelandais G, Devaux F, Jacq C (2003) Competitive promoter occupancy by two yeast paralogous transcription factors controlling the multidrug resistance phenomenon. J Biol Chem 278(52):52641–52650. doi:10.1074/jbc.M309580200
Ma M, Liu ZL (2010) Comparative transcriptome profiling analyses during the lag phase uncover YAP1, PDR1, PDR3, RPN4 and HSF1 as key regulatory genes in genomic adaptation to the lignocellulose derived inhibitor HMF for Saccharomyces cerevisiae. BMC Genomics 11:660. doi:10.1186/1471-2164-11-660
Ma M, Wang X, Zhang X, Zhao X (2013) Alcohol dehydrogenases from Scheffersomyces stipitis involved in the detoxification of aldehyde inhibitors derived from lignocellulosic biomass conversion. Appl Microbiol Biotechnol 97(18):8411–8425. doi:10.1007/s00253-013-5110-8
Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Lu S, Marchler GH, Song JS, Thanki N, Yamashita RA, Zhang D, Bryant SH (2013) CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res 41:D348–D352. doi:10.1093/nar/gks1243
Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH (2015) CDD: NCBI’s conserved domain database. Nucleic Acids Res 43:D222–D226. doi:10.1093/nar/gku1221
Moon J, Liu ZL (2015) Direct enzyme assay evidence confirms aldehyde reductase function of Ydr541cp and Ygl039wp from Saccharomyces cerevisiae. Yeast 32(4):399–407. doi:10.1002/yea.3067
Newstead S, Kim H, von Heijne G, Iwata S, Drew D (2007) High-throughput fluorescent-based optimization of eukaryotic membrane protein overexpression and purification in Saccharomyces cerevisiae. Proc Natl Acad Sci 104(35):13936–13941. doi:10.1073/pnas.0704546104
Nordling E, Jörnvall H, Persson B (2002) Medium-chain dehydrogenases/reductases (MDR): family characterizations including genome comparisons and active site modeling. Eur J Biochem 269(17):4267–4276. doi:10.1046/j.1432-1033.2002.03114.x
Peralba JM, Cederlund E, Crosas B, Moreno A, Julià P, Martinez SE, Persson B, Farrés J, Parés X, Jörnvall H (1999) Structural and enzymatic properties of a gastric NADP(H)-dependent and retinal-active alcohol dehydrogenase. J Biol Chem 274(37):26021–26026. doi:10.1074/jbc.274.37.26021
Persson B, Jeffery J, Jörnvall H (1991) Different segment similarities in long-chain dehydrogenases. Biochem Biophys Res Commun 177(1):218–223. doi:10.1016/0006-291X(91)91970-N
Persson B, Hedlund J, Jörnvall H (2008) Medium- and short-chain dehydrogenase/reductase gene and protein families: the MDR superfamily. Cell Mol Life Sci 65(24):3879–3894. doi:10.1007/s00018-008-8587-z
Schwartz MF, Jörnvall H (1976) Structural analyses of mutant and wild-type alcohol dehydrogenases from Drosophila melanogaster. Eur J Biochem 68:159–168. doi:10.1111/j.1432-1033.1976.tb10774.x
Sinicropi D, Cronin M, Liu M-L (2007) Gene expression profiling utilizing microarray technology and RT-PCR. In: Ferrari M, Ozkan M, Heller M (eds) BioMEMS and biomedical nanotechnology, Volume II: Micro/Nano technologies for genomics and proteomics. Springer-Verlag, Heidelberg, pp 23–46
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729. doi:10.1093/molbev/mst197
Teixeira MC, Monteiro PT, Guerreiro JF, Gonçalves JP, Mira NP, dos Santos SC, Cabrito TR, Palma M, Costa C, Francisco AP, Madeira SC, Oliveira AL, Freitas AT, Sá-Correia I (2014) The YEASTRACT database: an upgraded information system for the analysis of gene and genomic transcription regulation in Saccharomyces cerevisiae. Nucleic Acids Res 42:D161–D166. doi:10.1093/nar/gkt1015
Thatcher DR, Sawyer L (1980) Secondary-structure prediction from the sequence of Drosophila melanogaster (fruitfly) alcohol dehydrogenase. Biochem J 187(3):884–886. doi:10.1042/bj1870884
Van Dijken JP, Scheffers WA (1986) Redox balances in the metabolism of sugars by yeasts. FEMS Microb Rev 32(3–4):199–224. doi:10.1016/0378-1097(86)90291-0
Wang X, Ma M, Liu ZL, Xiang Q, Li X, Liu N, Zhang X (2016) GRE2 from Scheffersomyces stipitis as an aldehyde reductase contributes tolerance to aldehyde inhibitors derived from lignocellulosic biomass. Appl Microbiol Biotechnol 100(15):6671–6682. doi:10.1007/s00253-016-7445-4
Wu WS, Li WH (2008) Identifying gene regulatory modules of heat shock response in yeast. BMC Genomics 9:439. doi:10.1186/1471-2164-9-439
Yofe I, Weill U, Meurer M, Chuartzman S, Zalckvar E, Goldman O, Ben-Dor S, Schütze C, Wiedemann N, Knop M, Khmelinskii A, Schuldiner M (2016) One library to make them all: streamlining the creation of yeast libraries via a SWAp-Tag strategy. Nat Methods 13(4):371–378. doi:10.1038/nmeth.3795
Zhao X, Tang J, Wang X, Yang R, Zhang X, Gu Y, Li X, Ma M (2015) YNL134C from Saccharomyces cerevisiae encodes a novel protein with aldehyde reductase activity for detoxification of furfural derived from lignocellulosic biomass. Yeast 32(5):409–422. doi:10.1002/yea.3068
Acknowledgments
We thank Z. Lewis Liu, Bioenergy Research Unit, NCAUR-ARS, U.S. Department of Agriculture (Peoria, IL, USA) for providing the calibrated mRNA control mix as the reference mRNA for the qRT-PCR analyses. This work was supported by the National Natural Science Foundation of China (No. 31570086) and the Talent Introduction Fund of Sichuan Agricultural University (No. 01426100).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals by any of the authors.
Additional information
Han-Yu Wang and Di-Fan Xiao contributed equally to this work.
Electronic supplementary material
ESM 1
(PDF 391 kb)
Rights and permissions
About this article
Cite this article
Wang, HY., Xiao, DF., Zhou, C. et al. YLL056C from Saccharomyces cerevisiae encodes a novel protein with aldehyde reductase activity. Appl Microbiol Biotechnol 101, 4507–4520 (2017). https://doi.org/10.1007/s00253-017-8209-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8209-5