Abstract
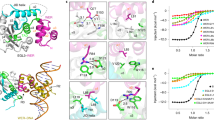
Regulatory factor MBF1 is highly conserved between species and has been described as a cofactor and transcription factor. In plants, several reports associate MBF1 with heat stress response. Nevertheless, the specific physical processes involved in the MBF1–DNA interaction are still far from clearly understood. We thus performed extensive molecular dynamics simulations of DNA with a homology-based modethel of the MBF1 protein. Based on recent experimental data, we proposed two B-DNA sequences, analyzing their interaction with our model of the Arabidopsis MBF1c protein (AtMBF1c) at three different temperatures: 293, 300, and 320 K, maintaining a constant pressure of 1 bar. The simulations suggest that MBF1 binds directly to the DNA, supporting the idea of its role as a transcription factor. We identified two different conformations of the MBF1 protein when bound, and characterized the specific groups of amino acids involved in the formation of the DNA–MBF1 complex. These regions of amino acids are bound mostly to the minor groove of DNA by the attraction of positively charged residues and the negatively charged backbone, but subject to the compatibility of shapes, much in the sense of a lock-and-key mechanism. We found that only with a sequence rich in CTAGA motifs at 300 K does MBF1 bind to DNA in the DNA-binding domain Cro/C1-type HTH predicted. In the rest of the systems tested, we observed non-specific DNA–MBF1 interactions. This study complements findings previously reported by others on the role of CTAGA as a DNA-binding element for MBF1c at a heat stress temperature.







Similar content being viewed by others
References
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindah E (2015) Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19–25. https://doi.org/10.1016/j.softx.2015.06.001
Alavilli H, Lee H, Park M, Lee BH (2017) Antarctic moss multiprotein bridging factor 1c overexpression in Arabidopsis resulted in enhanced tolerance to salt stress. Front Plant Sci 8:1206. https://doi.org/10.3389/fpls.2017.01206
Anderson W, Ohlendorf D, Takeda Y (1981) Structure of the cro repressor from bacteriophage and its interaction with dna. Nature 290:754–758. https://doi.org/10.1038/290754a0
Andrabi M, Hutchins AP, Miranda-Saavedra D, Kono H, Nussinov R, Mizuguchi K, Ahmad S (2017) Predicting conformational ensembles and genome-wide transcription factor binding sites from DNA sequences. Sci Rep 7(1):1–16. https://doi.org/10.1038/s41598-017-03199-6
Aravind L, Koonin EV (1999) DNA-binding proteins and evolution of transcription regulation in the archaea. Nucleic Acids Res 27(23):4658–4670
Battistini F, Hospital A, Buitrago D, Gallego D, Dans PD, Gelpí JL, Orozco M (2019) How B-DNA dynamics decipher sequence-selective protein recognition. J Mol Biol 431(19):3845–3859. https://doi.org/10.1016/j.jmb.2019.07.021
Berendsen HJ, Grigera JR, Straatsma TP (1987) The missing term in effective pair potentials. J Phys Chem 91(24):6269–6271. https://doi.org/10.1021/j100308a038
Bussi G, Donadio D, Parrinello M (2007) Canonical sampling through velocity rescaling. J Chem Phys 126(1):1–7. https://doi.org/10.1063/1.2408420
de Koning B, Blombach F, Wu H, Brouns SJJ, van der Oost J (2009) Role of multiprotein bridging factor 1 in archaea: bridging the domains? Biochem Soc Trans 37(Pt 1):52–7. https://doi.org/10.1042/BST0370052
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792–1797. https://doi.org/10.1093/nar/gkh340
El Hassan M, Calladine C (1998) Two distinct modes of protein-induced bending in dna11 edited by J. Karn. J Mol Biol 282(2):331–343. https://doi.org/10.1006/jmbi.1998.1994
Garvie CW, Wolberger C (2001) Recognition of specific dna sequences. Mol Cell 8:937–946. https://doi.org/10.1016/S1097-2765(01)00392-6
Hohenberg P, Kohn W (1964) Inhomogeneous electron gas. Phys Rev 136:B864–B871. https://doi.org/10.1103/PhysRev.136.B864
Hospital A, Faustino I, Collepardo-Guevara R, González C, Gelpí JL, Orozco M (2013) NAFlex: a web server for the study of nucleic acid flexibility. Nucleic Acids Res 41(Web Server issue):47–55. https://doi.org/10.1093/nar/gkt378
Ivani I, Dans PD, Noy A, Pérez A, Faustino I, Hospital A, Walther J, Andrio P, Goñi R, Balaceanu A, Portella G, Battistini F, Gelpí JL, González C, Vendruscolo M, Laughton CA, Harris SA, Case DA, Orozco M (2016) Parmbsc1: a refined force field for dna simulations. Nat Methods 13(1):55–58. https://doi.org/10.1038/nmeth.3658
Jaimes-Miranda F, Chávez Montes RA (2020) The plant MBF1 protein family: a bridge between stress and transcription. J Exp Bot 71(6):1782–1791. https://doi.org/10.1093/jxb/erz525
Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236. https://doi.org/10.1093/BIOINFORMATICS/BTU031
Kleinman L, Bylander DM (1982) Efficacious form for model pseudopotentials. Phys Rev Lett 48:1425–1428. https://doi.org/10.1103/PhysRevLett.48.1425
Kohn W, Sham LJ (1965) Self-consistent equations including exchange and correlation effects. Phys Rev 140:A1133–A1138. https://doi.org/10.1103/PhysRev.140.A1133
Li FQ, Ueda H, Hirose S (1994) Mediators of activation of fushi tarazu gene transcription by BmFTZ-F1. Mol Cell Biol 14(5):3013–21
Liao Q, Lüking M, Krüger DM, Deindl S, Elf J, Kasson PM, Lynn Kamerlin SC (2019) Long time-scale atomistic simulations of the structure and dynamics of transcription factor-DNA recognition. J Phys Chem B 123(17):3576–3590. https://doi.org/10.1021/acs.jpcb.8b12363
Lu X, Olson WK (2003) 3DNA: a software package for the analysis, rebuilding and visualization of three dimensional nucleic acid structures. Nucleic Acids Res 31(17):5108–5121. https://doi.org/10.1093/nar/gkg680
Marin-Gonzalez A, Vilhena JG, Moreno-Herrero F, Perez R (2019) DNA crookedness regulates DNA mechanical properties at short length scales. Phys Rev Lett 122(4):48102. https://doi.org/10.1103/PhysRevLett.122.048102
Matthews BW, Ohlendorf DH, Anderson WF, Fisher RG, Takeda Y (1983) Cro repressor protein and its interaction with DNA. Cold Spring Harb Symp Quant Biol 290:427–433. https://doi.org/10.1101/sqb.1983.047.01.050
Meza JC (2010) Steepest descent. Wiley Interdiscip Rev Comput Stat 2(6):719–722. https://doi.org/10.1002/wics.117
Oguey C, Foloppe N, Hartmann B (2010) Understanding the sequence-dependence of DNA groove dimensions: implications for DNA interactions. PLoS One 5(12):1–8. https://doi.org/10.1371/journal.pone.0015931
Parrinello M, Rahman A (1981) Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys 52(12):7182–7190. https://doi.org/10.1063/1.328693
Perdew JP, Ruzsinszky A, Csonka GI, Vydrov OA, Scuseria GE, Constantin LA, Zhou X, Burke K (2008) Restoring the density-gradient expansion for exchange in solids and surfaces. Phys Rev Lett 100:136406. https://doi.org/10.1103/PhysRevLett.100.136406
Perino M, Van Mierlo G, Karemaker ID, Van Genesen S, Vermeulen M, Marks H, Van Heeringen SJ, Veenstra GJC (2018) MTF2 recruits polycomb repressive complex 2 by helical-shape-selective DNA binding. Nat Genet 50(7):1002–1010. https://doi.org/10.1038/s41588-018-0134-8
Pnueli L, Hallak-Herr E, Rozenberg M, Cohen M, Goloubinoff P, Kaplan A, Mittler R (2002) Molecular and biochemical mechanisms associated with dormancy and drought tolerance in the desert legume Retama raetam. Plant J 31(3):319–330. https://doi.org/10.1046/j.1365-313X.2002.01364.x
Qin D, Wang F, Geng X, Zhang L, Yao Y, Ni Z, Peng H, Sun Q (2015) Overexpression of heat stress-responsive TaMBF1c, a wheat (Triticum aestivum L.) multiprotein bridging factor, confers heat tolerance in both yeast and rice. Plant Mol Biol 87(1–2):31–45. https://doi.org/10.1007/s11103-014-0259-9
Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS (2010) Origins of specificity in protein-DNA recognition. Annu Rev Biochem 79(1):233–269. https://doi.org/10.1146/annurev-biochem-060408-091030 (PMID: 20334529)
Samee MAH, Bruneau BG, Pollard KS (2019) A de novo shape motif discovery algorithm reveals preferences of transcription factors for DNA shape beyond sequence motifs. Cell Syst 8(1):27-42.e6. https://doi.org/10.1016/j.cels.2018.12.001
Sinha NK, Ordureau A, Best K, Saba JA, Zinshteyn B, Sundaramoorthy E, Fulzele A, Garshott DM, Denk T, Thoms M, Paulo JA, Harper JW, Bennett EJ, Beckmann R, Green R (2020) Edf1 coordinates cellular responses to ribosome collisions. eLife 9:e58828. https://doi.org/10.7554/eLife.58828
Soler JM, Artacho E, Gale JD, García A, Junquera J, Ordejón P, Sánchez-Portal D (2002) The SIESTA method for ab initio order-N materials simulation. J Phys Condens Matter 14(11):2745–2779. https://doi.org/10.1088/0953-8984/14/11/302
Suzuki N, Rizhsky L, Liang H, Shuman J, Shulaev V, Mittler R (2005) Enhanced tolerance to environmental stress in transgenic plants expressing the transcriptional coactivator multiprotein bridging factor 1c. Plant Physiol 139(3):1313–22. https://doi.org/10.1104/pp.105.070110
Suzuki N, Bajad S, Shuman J, Shulaev V, Mittler R (2008) The transcriptional co-activator MBF1c is a key regulator of thermotolerance in Arabidopsis thaliana. J Biol Chem 283(14):9269–75. https://doi.org/10.1074/jbc.M709187200
Suzuki N, Sejima H, Tam R, Schlauch K, Mittler R (2011) Identification of the MBF1 heat-response regulon of Arabidopsis thaliana. Plant J 66(5):844–851. https://doi.org/10.1111/j.1365-313X.2011.04550.x
Troullier N, Martins JL (1991) Efficient pseudopotentials for plane-wave calculations. Phys Rev B 43:1993–2006. https://doi.org/10.1103/PhysRevB.43.1993
Tsuda K, Yamazaki K (2004) Structure and expression analysis of three subtypes of Arabidopsis MBF1 genes. Biochim Biophys Acta (BBA) Gene Struct Express 1680(1):1–10. https://doi.org/10.1016/J.BBAEXP.2004.08.004
Uji T, Sato R, Mizuta H, Saga N (2013) Changes in membrane fluidity and phospholipase D activity are required for heat activation of PyMBF1 in Pyropia yezoensis (Rhodophyta). J Appl Phycol 25(6):1887–1893. https://doi.org/10.1007/s10811-013-0006-7
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, De Beer TA, Rempfer C, Bordoli L, Lepore R, Schwede T (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46(W1):W296–W303. https://doi.org/10.1093/nar/gky427
Wieczór M, Czub J (2017) How proteins bind to DNA: target discrimination and dynamic sequence search by the telomeric protein TRF1. Nucleic Acids Res 45(13):7643–7654. https://doi.org/10.1093/nar/gkx534
Wintjens R, Rooman M (1996) Structural classification of hth dna-binding domains and protein-dna interaction modes. J Mol Biol 262(2):294–313. https://doi.org/10.1006/jmbi.1996.0514
Yesudhas D, Anwar MA, Panneerselvam S, Kim HK, Choi S (2017) Evaluation of Sox2 binding affinities for distinct DNA patterns using steered molecular dynamics simulation. FEBS Open Bio 7(11):1750–1767. https://doi.org/10.1002/2211-5463.12316
Zhao S, Liu Q, Wang JX, Liao XZ, Guo H, Li CX, Zhang FF, Liao LS, Luo XM, Feng JX (2019) Differential transcriptomic profiling of filamentous fungus during solid-state and submerged fermentation and identification of an essential regulatory gene PoxMBF1 that directly regulated cellulase and xylanase gene expression. Biotechnol Biofuels 12(1):1–14. https://doi.org/10.1186/s13068-019-1445-4
Acknowledgements
The IPICYT’s National Supercomputing Center supported this research with the computational time Grants TKII-R2018-COV1, TKII-R2018-DSB1, TKII-R2018-FJM1, TKII-R2018-FLU1. FJM thanks financial support of the Consejo Nacional de Ciencia y Tecnología through Grant CB 2017-2018 A1-S-7679.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Salgado-Blanco, D., López-Urías, F., Ovando-Vázquez, C. et al. DNA–MBF1 study using molecular dynamics simulations. Eur Biophys J 50, 1055–1067 (2021). https://doi.org/10.1007/s00249-021-01565-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00249-021-01565-x